6LSS
 
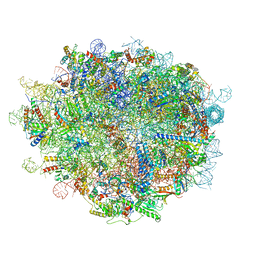 | | Cryo-EM structure of a pre-60S ribosomal subunit - state preA | | Descriptor: | 28S rRNA, 5S rRNA, 60S ribosomal protein L11, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-20 | | Release date: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6LSR
 
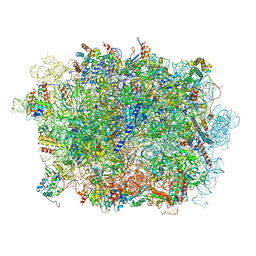 | | Cryo-EM structure of a pre-60S ribosomal subunit - state B | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-20 | | Release date: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
1ZXO
 
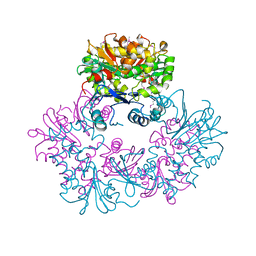 | | X-ray Crystal Structure of Protein Q8A1P1 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR25. | | Descriptor: | conserved hypothetical protein Q8A1P1 | | Authors: | Kuzin, A.P, Yong, W, Forouhar, F, Vorobiev, S, Xiao, R, Ma, C, Acton, T, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-06-08 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-Ray structure of the hypothetical protein Q8A1P1 at the resolution 3.2A. Northeast Structural Genomics Consortium target BtR25.
To be Published
|
|
2OAZ
 
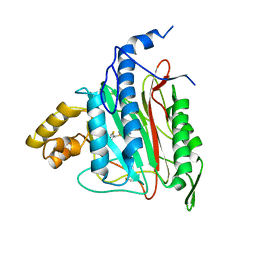 | | Human Methionine Aminopeptidase-2 Complexed with SB-587094 | | Descriptor: | COBALT (II) ION, N-(2-ISOPROPYLPHENYL)-3-[(2-THIENYLMETHYL)THIO]-1H-1,2,4-TRIAZOL-5-AMINE, human Methionine Amino Peptidase 2 | | Authors: | Marino Jr, J.P, Fisher, P.W, Hofmann, G.A, Kirkpatrick, R, Janson, C.A, Johnson, R.K, Ma, C, Mattern, M, Meek, T.D, Ryan, D, Schulz, C, Smith, W.W, Tew, D.G, Tomazek Jr, T.A, Veber, D.F, Xiong, W.C, Yamamoto, Y, Yamashita, K, Yang, G, Thompson, S.K. | | Deposit date: | 2006-12-18 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent inhibitors of methionine aminopeptidase-2 based on a 1,2,4-triazole pharmacophore.
J.Med.Chem., 50, 2007
|
|
3PFT
 
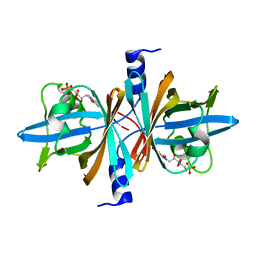 | | Crystal Structure of Untagged C54A Mutant Flavin Reductase (DszD) in Complex with FMN From Mycobacterium goodii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin reductase | | Authors: | Li, Q, Xu, P, Ma, C, Gu, L, Liu, X, Zhang, C, Li, N, Su, J, Li, B, Liu, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The flavin reductase DSZD from a desulfurizing mycobacterium goodii strain: systemic manipulation and investigation based on the crystal structure
To be Published
|
|
6A68
 
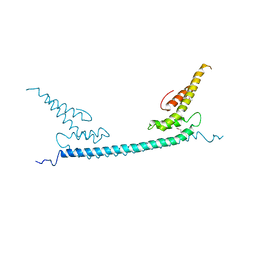 | | the crystal structure of rat calcium-dependent activator protein for secretion (CAPS) DAMH domain | | Descriptor: | Calcium-dependent secretion activator 1, POTASSIUM ION | | Authors: | Zhou, H, Wei, Z.Q, Yao, D.Q, Zhang, R.G, Ma, C. | | Deposit date: | 2018-06-26 | | Release date: | 2019-03-13 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structural and Functional Analysis of the CAPS SNARE-Binding Domain Required for SNARE Complex Formation and Exocytosis.
Cell Rep, 26, 2019
|
|
2AJA
 
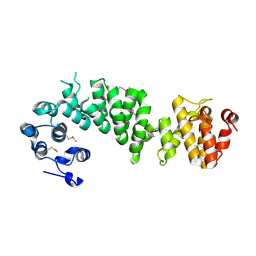 | | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR21. | | Descriptor: | ankyrin repeat family protein | | Authors: | Kuzin, A.P, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, C, Kellie, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila.
To be Published
|
|
6A30
 
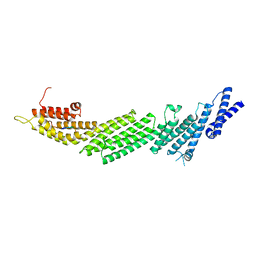 | | Crystal Structure of Munc13-1 MUN Domain and Synaptobrevin-2 Juxtamembrane Linker Region | | Descriptor: | Protein unc-13 homolog A, Synaptobrevin-2 juxtamembrane linker peptide | | Authors: | Wang, S, Li, Y, Gong, J.H, Ye, S, Yang, X.F, Zhang, R.G, Ma, C. | | Deposit date: | 2018-06-14 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly.
Nat Commun, 10, 2019
|
|
2K2U
 
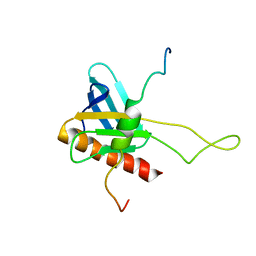 | | NMR Structure of the complex between Tfb1 subunit of TFIIH and the activation domain of VP16 | | Descriptor: | Alpha trans-inducing protein, RNA polymerase II transcription factor B subunit 1 | | Authors: | Langlois, C, Mas, C, Di Lello, P, Miller Jenkins, P.M, Legault, J, Omichinski, J.G. | | Deposit date: | 2008-04-11 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Complex between the Tfb1 Subunit of TFIIH and the Activation Domain of VP16: Structural Similarities between VP16 and p53.
J.Am.Chem.Soc., 130, 2008
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
7CFM
 
 | | Cryo-EM structure of the P395-bound GPBAR-Gs complex | | Descriptor: | 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
5GSQ
 
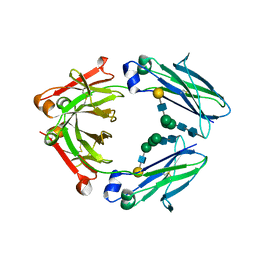 | | Crystal structure of IgG Fc with a homogeneous glycoform and Antibody-Dependent Cellular Cytotoxicity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, C.-L, Hsu, J.-C, Lin, C.-W, Wu, C.-Y, Wong, C.-H, Ma, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a Homogeneous IgG-Fc Glycoform with the N-Glycan Designed to Maximize the Antibody Dependent Cellular Cytotoxicity
ACS Chem. Biol., 12, 2017
|
|
3FWL
 
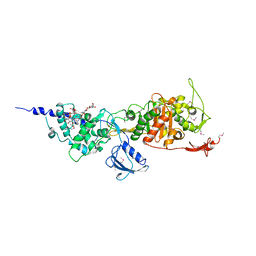 | | Crystal Structure of the Full-Length Transglycosylase PBP1b from Escherichia coli | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Sung, M.T, Lai, Y.T, Huang, C.Y, Chou, L.Y, Wong, C.H, Ma, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7CN9
 
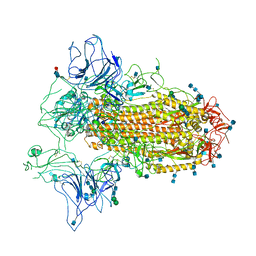 | | Cryo-EM structure of SARS-CoV-2 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ho, M, Chang, Y, Wang, C, Wu, Y, Huang, H, Chen, T, Lo, J.M, Chen, X, Ma, C. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Carbohydrate-Binding Protein from the Edible Lablab Beans Effectively Blocks the Infections of Influenza Viruses and SARS-CoV-2.
Cell Rep, 32, 2020
|
|
8ISI
 
 | | Photochromobilin-free form of Arabidopsis thaliana phytochrome A - apo-AtphyA | | Descriptor: | Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
3VMT
 
 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase in complex with a Lipid II analog | | Descriptor: | MAGNESIUM ION, Monofunctional glycosyltransferase, [(2R,3R,4R,5S,6R)-4-[(2R)-1-[[(2S)-1-[2-[2-[2-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]ethoxy]ethoxy]ethylamino]-1-oxidanylidene-propan-2-yl]amino]-1-oxidanylidene-propan-2-yl]oxy-3-acetamido-5-[(2S,3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-(hydroxymethyl)oxan-2-yl] [oxidanyl(3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaenoxy)phosphoryl] hydrogen phosphate | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VMR
 
 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase in complex with moenomycin | | Descriptor: | MOENOMYCIN, Monofunctional glycosyltransferase | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.688 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VMA
 
 | | Crystal Structure of the Full-Length Transglycosylase PBP1b from Escherichia coli | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Huang, C.Y, Sung, M.T, Lai, Y.T, Chou, L.Y, Shih, H.W, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.161 Å) | | Cite: | Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3VMS
 
 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase in complex with NBD-Lipid II | | Descriptor: | Monofunctional glycosyltransferase | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VMQ
 
 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase: Apoenzyme | | Descriptor: | MAGNESIUM ION, Monofunctional glycosyltransferase | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
3J3V
 
 | | Atomic model of the immature 50S subunit from Bacillus subtilis (state I-a) | | Descriptor: | 50S ribosomal protein L1, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Li, N, Guo, Q, Zhang, Y, Yuan, Y, Ma, C, Lei, J, Gao, N. | | Deposit date: | 2013-04-28 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.3 Å) | | Cite: | Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit
Nucleic Acids Res., 41, 2013
|
|
3J3W
 
 | | Atomic model of the immature 50S subunit from Bacillus subtilis (state II-a) | | Descriptor: | 50S ribosomal protein L1, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Li, N, Guo, Q, Zhang, Y, Yuan, Y, Ma, C, Lei, J, Gao, N. | | Deposit date: | 2013-04-28 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit
Nucleic Acids Res., 41, 2013
|
|
