2W84
 
 | | Structure of Pex14 in complex with Pex5 | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19.
EMBO J., 28, 2009
|
|
2W85
 
 | | Structure of Pex14 in complex with Pex19 | | Descriptor: | PEROXIN-19, PEROXISOMAL MEMBRANE ANCHOR PROTEIN PEX14 | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Competitive Interactions of Pex14 with the Import Receptors Pex5 and Pex19.
Embo J., 28, 2009
|
|
5U07
 
 | | CRISPR RNA-guided surveillance complex | | Descriptor: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | Authors: | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
2WIP
 
 | | STRUCTURE OF CDK2-CYCLIN A COMPLEXED WITH 8-ANILINO-1-METHYL-4,5-DIHYDRO- 1H-PYRAZOLO[4,3-H] QUINAZOLINE-3-CARBOXYLIC ACID | | Descriptor: | 1-methyl-8-(phenylamino)-4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline-3-carboxylic acid, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, ... | | Authors: | Brasca, M.G, Amboldi, N, Ballinari, D, Cameron, A.D, Casale, E, Cervi, G, Colombo, M, Colotta, F, Croci, V, Dalessio, R, Fiorentini, F, Isacchi, A, Mercurio, C, Moretti, W, Panzeri, A, Pastori, W, Pevarello, P, Quartieri, F, Roletto, F, Traquandi, G, Vianello, P, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of N,1,4,4-Tetramethyl-8-{[4-(4-Methylpiperazin-1-Yl)Phenyl]Amino}-4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline-3-Carboxamide (Pha-848125), a Potent, Orally Available Cyclin Dependent Kinase Inhibitor.
J.Med.Chem., 52, 2009
|
|
5U0A
 
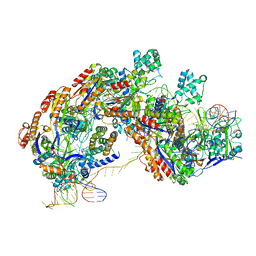 | | CRISPR RNA-guided surveillance complex | | Descriptor: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | Authors: | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
2XAG
 
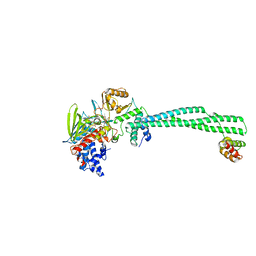 | | Crystal structure of LSD1-CoREST in complex with para-bromo-(-)-trans- 2-phenylcyclopropyl-1-amine | | Descriptor: | 3-(4-BROMOPHENYL)PROPANAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
3W9Z
 
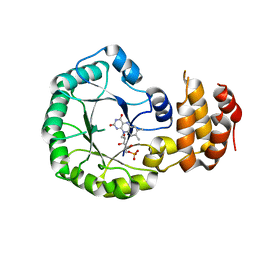 | | Crystal structure of DusC | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA-dihydrouridine synthase C | | Authors: | Chen, M, Yu, J, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2013-04-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydrouridine synthase C (DusC) from Escherichia coli
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2WMR
 
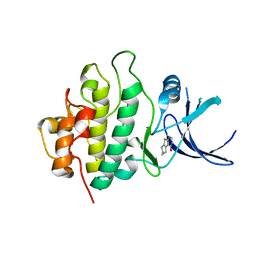 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 5,6,7,8-TETRAHYDRO[1]BENZOTHIENO[2,3-D]PYRIMIDIN-4(3H)-ONE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
2WOQ
 
 | | Porphobilinogen Synthase (HemB) in Complex with 5-acetamido-4- oxohexanoic acid (Alaremycin 2) | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALAREMYCIN 2, ... | | Authors: | Heinemann, I.U, Schulz, C, Schubert, W.-D, Heinz, D.W, Wang, Y.-G, Kobayashi, Y, Awa, Y, Wachi, M, Jahn, D, Jahn, M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the heme biosynthetic Pseudomonas aeruginosa porphobilinogen synthase in complex with the antibiotic alaremycin.
Antimicrob. Agents Chemother., 54, 2010
|
|
7JJE
 
 | |
2WJV
 
 | | Crystal structure of the complex between human nonsense mediated decay factors UPF1 and UPF2 | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 1, REGULATOR OF NONSENSE TRANSCRIPTS 2, SULFATE ION, ... | | Authors: | Clerici, M, Mourao, A, Gutsche, I, Gehring, N.H, Hentze, M.W, Kulozik, A, Kadlec, J, Sattler, M, Cusack, S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Unusual Bipartite Mode of Interaction between the Nonsense-Mediated Decay Factors, Upf1 and Upf2.
Embo J., 28, 2009
|
|
1KKU
 
 | | Crystal structure of nuclear human nicotinamide mononucleotide adenylyltransferase | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE | | Authors: | Garavaglia, S, D'Angelo, I, Emanuelli, M, Carnevali, F, Pierella, F, Magni, G, Rizzi, M. | | Deposit date: | 2001-12-10 | | Release date: | 2002-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of human NMN adenylyltransferase. A key nuclear enzyme for NAD homeostasis.
J.Biol.Chem., 277, 2002
|
|
7JJF
 
 | | Sarcin-ricin loop with modified residue. | | Descriptor: | MAGNESIUM ION, RNA/DNA (27-mer) | | Authors: | Harp, J.M, Pallan, P.S, Egli, M. | | Deposit date: | 2020-07-25 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Incorporating a Thiophosphate Modification into a Common RNA Tetraloop Motif Causes an Unanticipated Stability Boost.
Biochemistry, 59, 2020
|
|
3WGD
 
 | | Crystal structure of ERp46 Trx1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Inaba, K, Suzuki, M, Kojima, R. | | Deposit date: | 2013-08-04 | | Release date: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Radically different thioredoxin domain arrangement of ERp46, an efficient disulfide bond introducer of the mammalian PDI family
Structure, 22, 2014
|
|
1KHX
 
 | | Crystal structure of a phosphorylated Smad2 | | Descriptor: | Smad2 | | Authors: | Wu, J.-W, Hu, M, Chai, J, Seoane, J, Huse, M, Kyin, S, Muir, T.W, Fairman, R, Massague, J, Shi, Y. | | Deposit date: | 2001-12-01 | | Release date: | 2002-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling.
Mol.Cell, 8, 2001
|
|
3WGX
 
 | | Crystal structure of ERp46 Trx2 in a complex with Prx4 C-term | | Descriptor: | GLYCEROL, Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Inaba, K, Suzuki, M, Kojima, R. | | Deposit date: | 2013-08-13 | | Release date: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Radically different thioredoxin domain arrangement of ERp46, an efficient disulfide bond introducer of the mammalian PDI family
Structure, 22, 2014
|
|
2MJ2
 
 | | Structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix. | | Descriptor: | Agnoprotein | | Authors: | Coric, P, Saribas, S.A, Abou-Gharbia, M, Childers, W, White, M, Bouaziz, S, Safak, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix
J.Virol., 2014
|
|
5UGK
 
 | | Zinc-Binding Structure of a Catalytic Amyloid from Solid-State NMR Spectroscopy | | Descriptor: | ILE-HIS-VAL-HIS-LEU-GLN-ILE, ZINC ION | | Authors: | Lee, M, Wang, T, Makhlynets, O.V, Wu, Y, Polizzi, N, Wu, H, Gosavi, P.M, Korendovych, I.V, DeGrado, W.F, Hong, M. | | Deposit date: | 2017-01-09 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Zinc-binding structure of a catalytic amyloid from solid-state NMR.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2X31
 
 | | Modelling of the complex between subunits BchI and BchD of magnesium chelatase based on single-particle cryo-EM reconstruction at 7.5 ang | | Descriptor: | MAGNESIUM-CHELATASE 38 KDA SUBUNIT, MAGNESIUM-CHELATASE 60 KDA SUBUNIT | | Authors: | Lunqvist, J, Elmlund, H, Peterson Wulff, R, Berglund, L, Elmlund, D, Emanuelsson, C, Hebert, H, Willows, R.D, Hansson, M, Lindahl, M, Al-Karadaghi, S. | | Deposit date: | 2010-01-19 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | ATP-Induced Conformational Dynamics in the Aaa+ Motor Unit of Magnesium Chelatase.
Structure, 18, 2010
|
|
3WUL
 
 | |
5TZS
 
 | | Architecture of the yeast small subunit processome | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 3' domain-associated, ... | | Authors: | Chaker-Margot, M, Barandun, J, Hunziker, M, Klinge, S. | | Deposit date: | 2016-11-22 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Architecture of the yeast small subunit processome.
Science, 355, 2017
|
|
3AJD
 
 | | Crystal structure of ATRM4 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Putative methyltransferase MJ0026 | | Authors: | Hirano, M, Kuratani, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-01 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of Methanocaldococcus jannaschii Trm4 complexed with sinefungin.
J.Mol.Biol., 401, 2010
|
|
1JDE
 
 | | K22A mutant of pyruvate, phosphate dikinase | | Descriptor: | PYRUVATE, PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Ye, D, Wei, M, McGuire, M, Huang, K, Kapadia, G, Herzberg, O, Martin, B.M, Dunaway-Mariano, D. | | Deposit date: | 2001-06-13 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of the catalytic site within the ATP-grasp domain of Clostridium symbiosum pyruvate phosphate dikinase.
J.Biol.Chem., 276, 2001
|
|
4P6T
 
 | | Crystal Structure of tyrosinase from Bacillus megaterium with p-tyrosol in the active site | | Descriptor: | 4-(2-hydroxyethyl)phenol, COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2014-03-25 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins.
Nat Commun, 5, 2014
|
|
3WPX
 
 | | Structure of PomBc4, a periplasmic fragment of PomB from Vibrio alginolyticus | | Descriptor: | PomB | | Authors: | Takao, M, Sakuma, M, Zhu, S, Homma, M, Kojima, S, Imada, K. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational change in the periplasmic region of the flagellar stator coupled with the assembly around the rotor
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
