7LY3
 
 | |
6H5C
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 1 | | Descriptor: | (1~{S},3~{R},4~{S},5~{R})-3-methyl-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic Acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 9, 2019
|
|
4LAZ
 
 | | Crystal structure of human AR complexed with NADP+ and {5-chloro-2-[(4-iodobenzyl)carbamoyl]phenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {5-chloro-2-[(4-iodobenzyl)carbamoyl]phenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-20 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
6GZA
 
 | | Structure of murine leukemia virus capsid C-terminal domain | | Descriptor: | COBALT (II) ION, Capsid protein | | Authors: | Qu, K, Dolezal, M, Schur, F.K.M, Murciano, B, Rumlova, M, Ruml, T, Briggs, J.A.G. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and architecture of immature and mature murine leukemia virus capsids.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5N13
 
 | | Second Bromodomain (BD2) from Candida albicans Bdf1 in the unbound form | | Descriptor: | Bromodomain-containing factor 1, GLYCEROL | | Authors: | Mietton, F, Ferri, E, Champlebouxm, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5N17
 
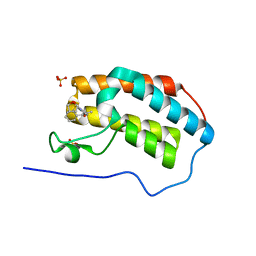 | | First Bromodomain (BD1) from Candida albicans Bdf1 bound to a dibenzothiazepinone (compound 3) | | Descriptor: | (2~{S})-~{N}-(5-methyl-6-oxidanylidene-benzo[b][1,4]benzothiazepin-2-yl)oxolane-2-carboxamide, Bromodomain-containing factor 1, SULFATE ION | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvon, M, d'Enfer, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5E6X
 
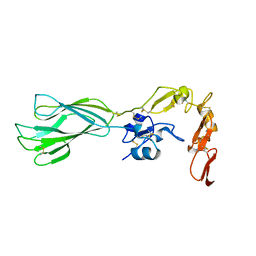 | |
5M3U
 
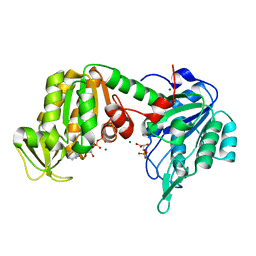 | | The X-ray structure of human V216F phosphoglycerate kinase 1 mutant | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ilari, A, Fiorillo, A, Cipollone, A, Petrosino, M. | | Deposit date: | 2016-10-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
5E78
 
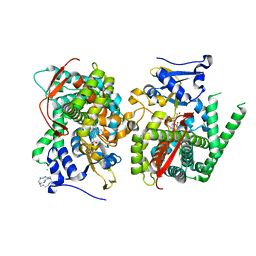 | | Crystal structure of P450 BM3 heme domain variant complexed with Co(III)Sep | | Descriptor: | 1,3,6,8,10,13,16,19-octaazabicyclo[6.6.6]icosane, Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, ... | | Authors: | Panneerselvm, S, Shehzad, A, Bocola, M, Mueller-Dieckmann, J, Schwaneberg, U. | | Deposit date: | 2015-10-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic insights into a cobalt (III) sepulchrate based alternative cofactor system of P450 BM3 monooxygenase.
Biochim. Biophys. Acta, 1866, 2018
|
|
6H04
 
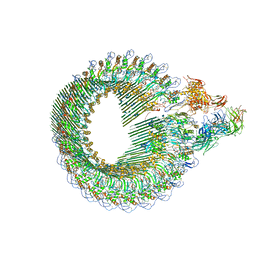 | | Closed conformation of the Membrane Attack Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5,Complement C5, ... | | Authors: | Menny, A, Serna, M, Boyd, C.M, Gardner, S, Joseph, A.P, Topf, M, Bubeck, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | CryoEM reveals how the complement membrane attack complex ruptures lipid bilayers.
Nat Commun, 9, 2018
|
|
6H5J
 
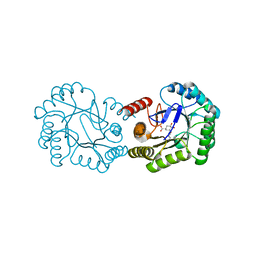 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 4 | | Descriptor: | (3~{R})-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Otero, J.M, Maneiro, M, Lence, E, Sanz-Gaitero, M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 6, 2019
|
|
5DL1
 
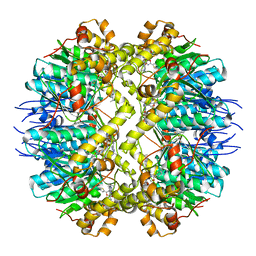 | | ClpP from Staphylococcus aureus in complex with AV145 | | Descriptor: | 1-(propan-2-yl)-N-{[2-(thiophen-2-yl)-1,3-oxazol-4-yl]methyl}-1H-pyrazolo[3,4-b]pyridine-5-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Vielberg, M.-T, Groll, M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Reversible Inhibitors Arrest ClpP in a Defined Conformational State that Can Be Revoked by ClpX Association.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6GZ5
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-3 (TI-POST-3) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
5ED6
 
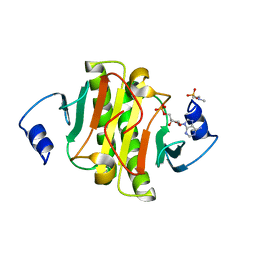 | | crystal structure of human Hint1 H114A mutant complexing with ATP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2015-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
5LS2
 
 | | Receptor mediated chitin perception in legumes is functionally seperable from Nod factor perception | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase, SULFATE ION | | Authors: | Bozsoki, Z, Cheng, J, Feng, F, Gysel, K, Andersen, K.R, Oldroyd, G, Blaise, M, Radutoiu, S, Stougaard, J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6GXO
 
 | | Cryo-EM structure of a rotated E. coli 70S ribosome in complex with RF3-GDPCP, RF1(GAQ) and P/E-tRNA (State IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
5LWW
 
 | | Crystal structure of a laccase-like multicopper oxidase McoG from Aspergillus niger bound to zinc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Ferraroni, M, Briganti, F, Tamayo-Ramos, J.A, van Berkel, W.J.H, Westphal, A.H. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and function of Aspergillus niger laccase McoG
Biocatalysis, 2017
|
|
1HKU
 
 | | CtBP/BARS: a dual-function protein involved in transcription corepression and Golgi membrane fission | | Descriptor: | C-TERMINAL BINDING PROTEIN 3, FORMIC ACID, GLYCEROL, ... | | Authors: | Nardini, M, Spano, S, Cericola, C, Pesce, A, Massaro, A, Millo, E, Luini, A, Corda, D, Bolognesi, M. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-19 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ctbp/Bars: A Dual-Function Protein Involved in Transcription Co-Repression and Golgi Membrane Fission
Embo J., 22, 2003
|
|
6GGJ
 
 | | Crystal structure of CotB2 in complex with alendronate | | Descriptor: | 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, Cyclooctat-9-en-7-ol synthase, MAGNESIUM ION | | Authors: | Driller, R, Janke, S, Fuchs, M, Warner, E, Mhashal, A.R, Major, D.T, Christmann, M, Brueck, T, Loll, B. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards a comprehensive understanding of the structural dynamics of a bacterial diterpene synthase during catalysis.
Nat Commun, 9, 2018
|
|
5LWN
 
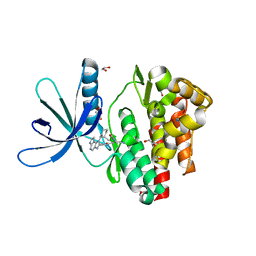 | | Crystal structure of JAK3 in complex with Compound 5 (FM409) | | Descriptor: | (2~{S})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]propanamide, (~{Z})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]prop-2-enamide, 1,2-ETHANEDIOL, ... | | Authors: | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
3MH0
 
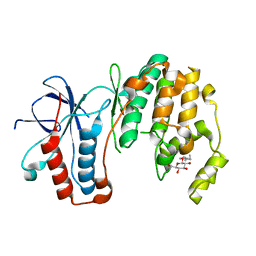 | | Mutagenesis of p38 MAP Kinase eshtablishes key roles of Phe169 in function and structural dynamics and reveals a novel DFG-out state | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Namboodiri, H.V, Karpusas, M, Bukhtiyarova, M, Springman, E.B. | | Deposit date: | 2010-04-07 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis of p38alpha MAP kinase establishes key roles of Phe169 in function and structural dynamics and reveals a novel DFG-OUT state.
Biochemistry, 46, 2007
|
|
4L3P
 
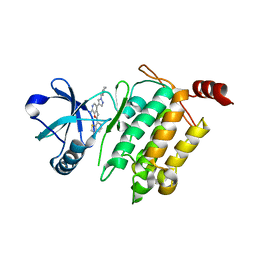 | | Crystal Structure of 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine bound to TAK1-TAB1 | | Descriptor: | 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Steinbacher, S, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-06 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and optimization of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5LYG
 
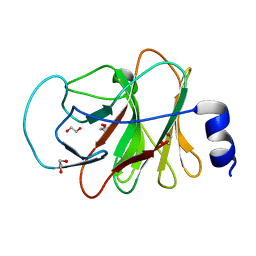 | | CRYSTAL STRUCTURE OF INTRACELLULAR B30.2 DOMAIN OF BTN3A1 BOUND TO MALONATE | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, MALONATE ION | | Authors: | Mohammed, F, Baker, A.T, Salim, M, Willcox, B.E. | | Deposit date: | 2016-09-28 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | BTN3A1 Discriminates gamma delta T Cell Phosphoantigens from Nonantigenic Small Molecules via a Conformational Sensor in Its B30.2 Domain.
ACS Chem. Biol., 12, 2017
|
|
3MHK
 
 | | Human tankyrase 2 - catalytic PARP domain in complex with 2-(2-pyridyl)-7,8-dihydro-5h-thiino[4,3-d]pyrimidin-4-ol | | Descriptor: | 2-pyridin-2-yl-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, GLYCEROL, Tankyrase-2, ... | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Markova, N, Moche, M, Nordlund, P, Nyman, T, Persson, C, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
3EJI
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36K at cryogenic temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, Thermonuclease | | Authors: | Khangulov, V.S, Schlessman, J.L, Garcia-Moreno, E.B, Benning, M, Isom, D. | | Deposit date: | 2008-09-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Consequences of the Ionization of Lysines at 25 Internal Positions in a Protein
To be Published
|
|
