4V9S
 
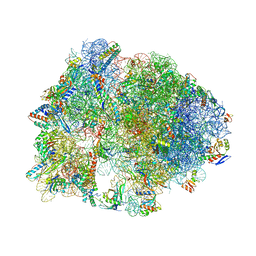 | | Crystal structure of antibiotic GE82832 bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4W6Y
 
 | | Co-complex structure of the lectin domain of F18 fimbrial adhesin FedF with inhibitory nanobody NbFedF9 | | Descriptor: | F18 fimbrial adhesin AC, Nanobody NbFedF9, SULFATE ION | | Authors: | Moonens, K, De Kerpel, M, Coddens, A, Cox, E, Pardon, E, Remaut, H, De Greve, H. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Nanobody Mediated Inhibition of Attachment of F18 Fimbriae Expressing Escherichia coli.
Plos One, 9, 2014
|
|
7LVY
 
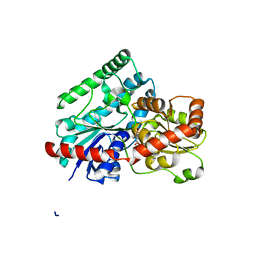 | | Crystal Structure of Tetur04g02350 | | Descriptor: | UDP-glycosyltransferase 203A2, URIDINE-5'-DIPHOSPHATE, beta-D-glucopyranose | | Authors: | Danehsian, L, Kluza, A, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2021-02-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Tetur04g02350
To Be Published
|
|
3KKE
 
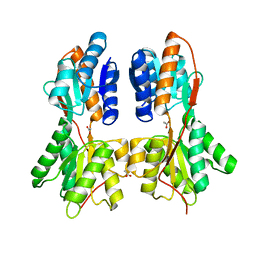 | | Crystal structure of a LacI family transcriptional regulator from Mycobacterium smegmatis | | Descriptor: | ACETATE ION, LacI family Transcriptional regulator | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Do, J, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-05 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a LacI family transcriptional regulator from Mycobacterium smegmatis
To be Published
|
|
4V16
 
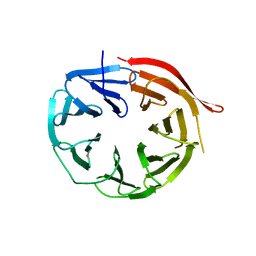 | | KlHsv2 with loop 6CD replaced by a Gly-Ser linker | | Descriptor: | SVP1-LIKE PROTEIN 2 | | Authors: | Busse, R.A, Scacioc, A, Krick, R, Perez-Lara, A, Thumm, M, Kuhnel, K. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of Proppin-Phosphoinositide Binding and Role of Loop 6Cd in Proppin-Membrane Binding.
Biophys.J., 108, 2015
|
|
7QWN
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
3KM0
 
 | | 17betaHSD1 in complex with 3beta-diol | | Descriptor: | 5-ALPHA-ANDROSTANE-3-BETA,17BETA-DIOL, Estradiol 17-beta-dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mazumdar, M, Lin, S.-X. | | Deposit date: | 2009-11-09 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of sex-steroid translation
To be Published
|
|
4WCJ
 
 | | Structure of IcaB from Ammonifex degensii | | Descriptor: | CHLORIDE ION, Polysaccharide deacetylase, ZINC ION | | Authors: | Little, D.J, Bamford, N.C, Pokrovskaya, V, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the De-N-acetylation of Poly-beta-1,6-N-acetyl-d-glucosamine in Gram-positive Bacteria.
J.Biol.Chem., 289, 2014
|
|
3KDN
 
 | | Crystal structure of Type III Rubisco SP4 mutant complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
4UXN
 
 | | LSD1(KDM1A)-CoREST in complex with Z-Pro derivative of MC2580 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | Authors: | Rodriguez, V, Valente, S, Stazi, G, Lucidi, A, Mercurio, C, Vianello, P, Ciossani, G, Mattevi, A, Botrugno, O.A, Dessanti, P, Minucci, S, Varasi, M, Mai, A. | | Deposit date: | 2014-08-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Pyrrole- and Indole-Containing Tranylcypromine Derivatives as Novel Lysine-Specific Demethylase 1 Inhibitors Active on Cancer Cells
Chemmedchem, 6, 2015
|
|
4UY4
 
 | | 1.86 A structure of human Spindlin-4 protein in complex with histone H3K4me3 peptide | | Descriptor: | GLYCEROL, HISTONE H3K4ME3, SPINDLIN-4 | | Authors: | Talon, R, Gileadi, C, Johansson, C, Burgess-Brown, N, Shrestha, L, von Delft, F, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | 1.86 A Structure of Human Spindlin-4 Protein in Complex with Histone H3K4Me3 Peptide
To be Published
|
|
4WEI
 
 | | Crystal structure of the F4 fimbrial adhesin FaeG in complex with lactose | | Descriptor: | K88 fimbrial protein AD, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Van den Broeck, I, De Kerpel, M, Deboeck, F, Raymaekers, H, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Insight into the Carbohydrate Receptor Binding of F4 Fimbriae-producing Enterotoxigenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
7LUG
 
 | | Crystal structure of the pnRFP B30Y mutant | | Descriptor: | PHOSPHATE ION, Red Fluorescent pnRFP B30Y mutant | | Authors: | Huang, M, Ng, H.L, Pang, Y, Zhang, S, Fan, Y, Yeh, H, Xiong, Y, Li, X, Ai, H. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development, Characterization, and Structural Analysis of a Genetically Encoded Red Fluorescent Peroxynitrite Biosensor
To Be Published
|
|
4USM
 
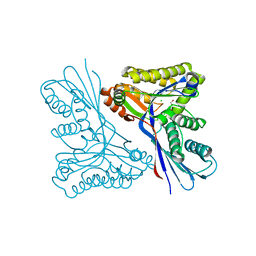 | | WcbL complex with glycerol bound to sugar site | | Descriptor: | CHLORIDE ION, GLYCEROL, PUTATIVE SUGAR KINASE | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
4UYU
 
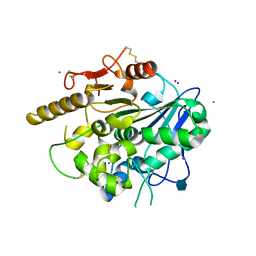 | |
3L02
 
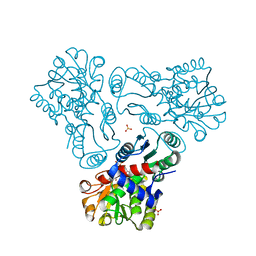 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase E92A mutant complexed with carbamyl phosphate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
Protein Sci., 16, 2007
|
|
4V19
 
 | | Structure of the large subunit of the mammalian mitoribosome, part 1 of 2 | | Descriptor: | MAGNESIUM ION, MITORIBOSOMAL 16S RRNA, MITORIBOSOMAL CP TRNA, ... | | Authors: | Greber, B.J, Boehringer, D, Leibundgut, M, Bieri, P, Leitner, A, Schmitz, N, Aebersold, R, Ban, N. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Complete Structure of the Large Subunit of the Mammalian Mitochondrial Ribosome
Nature, 515, 2014
|
|
4UVY
 
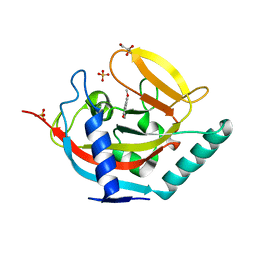 | | Crystal structure of human tankyrase 2 in complex with 3-(4- chlorophenyl)-5-methoxy-1,2- dihydroisoquinolin-1-one | | Descriptor: | 3-(4-chlorophenyl)-5-methoxyisoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
7LQO
 
 | | Crystal structure of a genetically encoded red fluorescent peroxynitrite biosensor, pnRFP | | Descriptor: | PHOSPHATE ION, red fluorescent peroxynitrite biosensor pnRFP | | Authors: | Huang, M, Ng, H.L, Pang, Y, Zhang, S, Fan, Y, Yeh, H, Xiong, Y, Li, X, Ai, H. | | Deposit date: | 2021-02-14 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development, Characterization, and Structural Analysis of a Genetically Encoded Red Fluorescent Peroxynitrite Biosensor
To Be Published
|
|
4US1
 
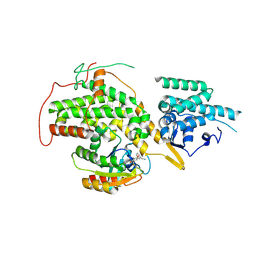 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | (3S)-3-[3-(aminomethyl)phenyl]-1-ethylpyrrolidine-2,5-dione, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
3L0O
 
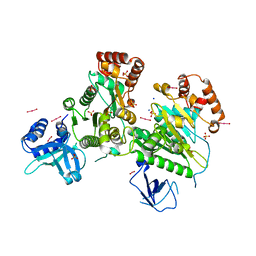 | | Structure of RNA-free Rho transcription termination factor from Thermotoga maritima | | Descriptor: | SODIUM ION, SULFATE ION, Transcription termination factor rho, ... | | Authors: | Canals, A, Uson, I, Coll, M. | | Deposit date: | 2009-12-10 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structure of RNA-Free Rho Termination Factor Indicates a Dynamic Mechanism of Transcript Capture
J.Mol.Biol., 400, 2010
|
|
4USZ
 
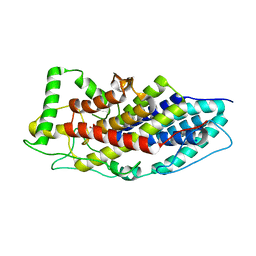 | | Crystal structure of the first bacterial vanadium dependant iodoperoxidase | | Descriptor: | SODIUM ION, VANADATE ION, VANADIUM-DEPENDENT HALOPEROXIDASE | | Authors: | Rebuffet, E, Delage, L, Fournier, J.B, Rzonca, J, Potin, P, Michel, G, Czjzek, M, Leblanc, C. | | Deposit date: | 2014-07-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Bacterial Vanadium Iodoperoxidase from the Marine Flavobacteriaceae Zobellia Galactanivorans Reveals Novel Molecular and Evolutionary Features of Halide Specificity in This Enzyme Family.
Appl.Environ.Microbiol., 80, 2014
|
|
4URN
 
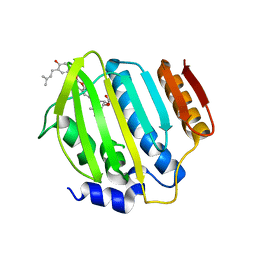 | | Crystal Structure of Staph ParE 24kDa in complex with Novobiocin | | Descriptor: | DNA TOPOISOMERASE IV, B SUBUNIT, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4V08
 
 | | Inhibited dimeric pseudorabies virus protease pUL26N at 2 A resolution | | Descriptor: | CHLORIDE ION, DIISOPROPYL PHOSPHONATE, MAGNESIUM ION, ... | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-09-11 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease
Plos Pathog., 11, 2015
|
|
3KEF
 
 | | Crystal structure of IspH:DMAPP-complex | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, DIMETHYLALLYL DIPHOSPHATE, FE3-S4 CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
