7MXC
 
 | | PRMT5:MEP50 complexed with adenosine | | Descriptor: | ADENOSINE, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
5AH0
 
 | | STRUCTURE OF LIPASE 1 FROM PELOSINUS FERMENTANS | | Descriptor: | DI(HYDROXYETHYL)ETHER, LIPASE, POTASSIUM ION, ... | | Authors: | Hromic, A, Gruber, K, Biundo, A, Ribitsch, D, Quartinello, F, Perz, V, Arrell, M.S, Kalman, F, Guebitz, G.M. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of a Poly(Butylene Adipate-Co-Terephthalate)-Hydrolyzing Lipase from Pelosinus Fermentans.
Appl.Microbiol.Biotechnol., 100, 2016
|
|
7MXA
 
 | | PRMT5:MEP50 complexed with inhibitor PF-06855800 | | Descriptor: | 7-[(5R)-5-C-(4-chloro-3-fluorophenyl)-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
7MXG
 
 | | PRMT5(M420T mutant):MEP50 complexed with inhibitor PF-06855800 | | Descriptor: | 7-[(5R)-5-C-(4-chloro-3-fluorophenyl)-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | McTigue, M, Deng, Y.L, Liu, W, Brooun, A. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.
Mol.Cancer Ther., 21, 2022
|
|
4GDP
 
 | | Yeast polyamine oxidase FMS1, N195A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1, TETRAETHYLENE GLYCOL | | Authors: | Taylor, A.B, Adachi, M.S, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2012-08-01 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9998 Å) | | Cite: | Mechanistic and Structural Analyses of the Roles of Active Site Residues in Yeast Polyamine Oxidase Fms1: Characterization of the N195A and D94N Enzymes.
Biochemistry, 51, 2012
|
|
4ECH
 
 | | Yeast Polyamine Oxidase FMS1, H67Q Mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1 | | Authors: | Taylor, A.B, Adachi, M.S, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2012-03-26 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic and structural analyses of the role of his67 in the yeast polyamine oxidase fms1.
Biochemistry, 51, 2012
|
|
4ERZ
 
 | | X-ray structure of WDR5-MLL4 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4Y48
 
 | | Endothiapepsin in complex with fragment B50 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4E8Z
 
 | | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | | Descriptor: | D-beta-D-heptose 7-phosphate kinase, POTASSIUM ION, {[2-({[5-(2,6-dichlorophenyl)-1,2,4-triazin-3-yl]amino}methyl)-1,3-benzothiazol-5-yl]oxy}acetic acid | | Authors: | Lee, T.-W, Verhey, T.B, Junop, M.S. | | Deposit date: | 2012-03-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4ZTT
 
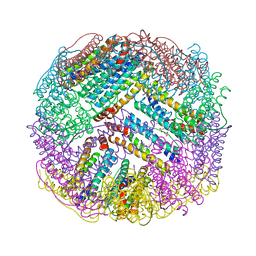 | | Crystal structures of ferritin mutants reveal diferric-peroxo intermediates | | Descriptor: | FE (II) ION, FE (III) ION, GLYCEROL, ... | | Authors: | Kim, S, Park, Y.H, Jung, S.W, Seok, J.H, Chung, Y.B, Lee, D.B, Gowda, G, Lee, J.H, Han, H.R, Cho, A.E, Lee, C, Chung, M.S, Kim, K.H. | | Deposit date: | 2015-05-15 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
6EL5
 
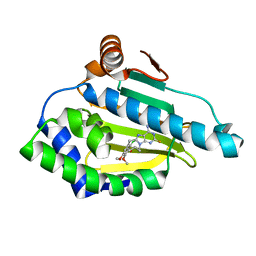 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-2-FLUORO-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, Heat shock protein HSP 90-alpha | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
4GQI
 
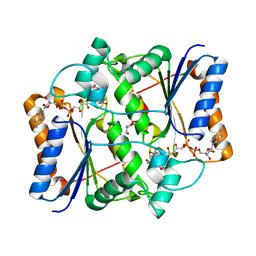 | | Synthesis of novel MT3 receptor ligands via unusual Knoevenagel condensation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N-{(3R)-3-[2-(acetylamino)ethyl]-2-oxo-2,3-dihydro-1H-indol-5-yl}acetamide, ... | | Authors: | Volkova, M.S, Jensen, K.C, Lozinskaya, N.A, Sosonyuk, S.E, Proskurnina, M.V, Mesecar, A.D, Zefirov, N.S. | | Deposit date: | 2012-08-23 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Synthesis of novel МТ3 receptor ligands via an unusual Knoevenagel condensation.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4EPQ
 
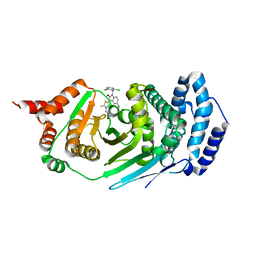 | |
4EPP
 
 | |
3GYQ
 
 | |
4ERY
 
 | | X-ray structure of WDR5-MLL3 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
6EI5
 
 | |
1BB0
 
 | | THROMBIN INHIBITORS WITH RIGID TRIPEPTIDYL ALDEHYDES | | Descriptor: | 2-{(3S)-3-[(benzylsulfonyl)amino]-2-oxopiperidin-1-yl}-N-{(2S)-1-[(3R)-1-carbamimidoylpiperidin-3-yl]-3-oxopropan-2-yl}acetamide, HIRUGEN, SODIUM ION, ... | | Authors: | Krishnan, R, Zhang, E, Hakansson, K, Arni, R.K, Tulinsky, A, Lim-Wilby, M.S.L, Levy, O.E, Semple, J.E, Brunck, T.K. | | Deposit date: | 1998-04-28 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes.
Biochemistry, 37, 1998
|
|
4Y4D
 
 | | Endothiapepsin in complex with fragment B51 | | Descriptor: | ACETATE ION, CAFFEINE, DIMETHYL SULFOXIDE, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4EXW
 
 | |
3IXY
 
 | | The pseudo-atomic structure of dengue immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
4GVY
 
 | | Crystal structure of arginine kinase in complex with L-citrulline and MgADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, CITRULLINE, ... | | Authors: | Clark, S.A, Davulcu, O, Chapman, M.S. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Crystal structures of arginine kinase in complex with ADP, nitrate, and various phosphagen analogs.
Biochem.Biophys.Res.Commun., 427, 2012
|
|
5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X1F
 
 | | CO bound cytochrome c oxidase without pump laser irradiation at 278K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
