1XB9
 
 | | The structure and function of Xenopus NO38-core, a histone chaperone in the nucleolus | | Descriptor: | Nucleophosmin | | Authors: | Namboodiri, V.M, Akey, I.V, Schmidt-Zachmann, M.S, Head, J.F, Akey, C.W. | | Deposit date: | 2004-08-30 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure and Function of Xenopus NO38-Core, a Histone Chaperone in the Nucleolus.
Structure, 12, 2004
|
|
3V3X
 
 | | Nitroxide Spin Labels in Protein GB1: N8/K28 Double Mutant | | Descriptor: | ACETATE ION, GLYCEROL, Immunoglobulin G-binding protein G, ... | | Authors: | Cunningham, T.F, McGoff, M.S, Sengupta, I, Jaroniec, C.P, Horne, W.S, Saxena, S.K. | | Deposit date: | 2011-12-14 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a protein spin-label in a solvent-exposed beta-sheet and comparison with DEER spectroscopy.
Biochemistry, 51, 2012
|
|
1W8D
 
 | | Binary structure of human DECR. | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, MITOCHONDRIAL PRECURSOR, GLYCEROL, ... | | Authors: | Alphey, M.S, Byres, E, Hunter, W.N. | | Deposit date: | 2004-09-20 | | Release date: | 2004-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Reactivity of Human Mitochondrial 2,4-Dienoyl-Coa Reductase: Enzyme-Ligand Interactions in a Distinctive Short-Chain Reductase Active Site
J.Biol.Chem., 280, 2005
|
|
3M0O
 
 | |
3M55
 
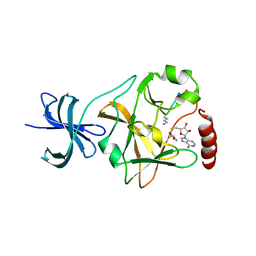 | | SET7/9 Y305F in complex with TAF10-K189me1 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10 peptide | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M53
 
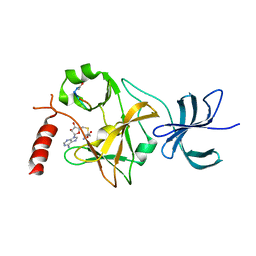 | | SET7/9 in complex with TAF10 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10 peptide | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3UXM
 
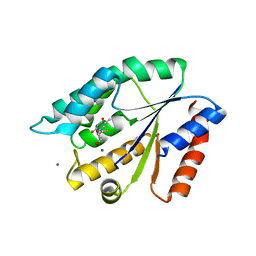 | | Structure Guided Development of Novel Thymidine Mimetics targeting Pseudomonas aeruginosa Thymidylate Kinase: from Hit to Lead Generation | | Descriptor: | 5'-deoxy-5'-fluorothymidine, MAGNESIUM ION, Thymidylate kinase | | Authors: | Choi, J.Y, Plummer, M.S, Starr, J, Desbonnet, C.R, Soutter, H.H, Chang, J, Miller, J.R, Dillman, K, Miller, A.A, Roush, W.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure guided development of novel thymidine mimetics targeting Pseudomonas aeruginosa thymidylate kinase: from hit to lead generation.
J.Med.Chem., 55, 2012
|
|
6G3B
 
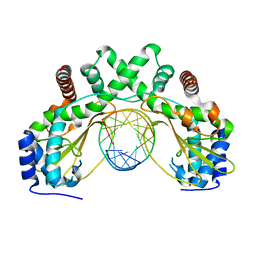 | | AvaII restriction endonuclease in complex with an RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*GP*GP*TP*CP*CP*TP*A)-3'), RNA (5'-R(P*GP*UP*AP*GP*GP*AP*CP*CP*AP*UP*G)-3'), Type II site-specific deoxyribonuclease | | Authors: | Kisiala, M, Kowalska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2018-03-24 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation
Nucleic Acids Res., 2020
|
|
2WOV
 
 | | Trypanosoma brucei trypanothione reductase with bound NADP. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-30 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WPC
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00073357) | | Descriptor: | (4S)-6-CHLORO-3-{2-[4-(FURAN-2-YLCARBONYL)PIPERAZIN-1-YL]ETHYL}-2-METHYL-4-PHENYL-3,4-DIHYDROQUINAZOLINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-05 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
7N12
 
 | | Crystal structure of the M. abscessus LeuRS editing domain in complex with epetraborole-AMP adduct | | Descriptor: | Leucine--tRNA ligase, SULFATE ION, [(1S,5R,6R,7'S,8R)-7'-(aminomethyl)-6-(6-aminopurin-9-yl)-2'-(3-oxidanylpropoxy)spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,9'-8-oxa-9-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-8-yl]methyl dihydrogen phosphate | | Authors: | Kalthoff, E, Schmeing, M. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Efficacy of epetraborole against Mycobacterium abscessus is increased with norvaline.
Plos Pathog., 17, 2021
|
|
7N11
 
 | |
1MCZ
 
 | | BENZOYLFORMATE DECARBOXYLASE FROM PSEUDOMONAS PUTIDA COMPLEXED WITH AN INHIBITOR, R-MANDELATE | | Descriptor: | (R)-MANDELIC ACID, BENZOYLFORMATE DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Polovnikova, E.S, Bera, A.K, Hasson, M.S. | | Deposit date: | 2002-08-06 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Kinetic Analysis of Catalysis by a Thiamin
Diphosphate-Dependent Enzyme, Benzoylformate Decarboxylase
Biochemistry, 42, 2003
|
|
2WPF
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00085762) | | Descriptor: | 3-[(4S)-6-CHLORO-2-METHYL-4-(4-METHYLPHENYL)QUINAZOLIN-3(4H)-YL]-N,N-DIMETHYLPROPAN-1-AMINE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
8DRX
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp10-nsp11 (C10) cut site sequence (form 2) | | Descriptor: | Fusion protein of 3C-like proteinase nsp5 and nsp10-nsp11 (C10) cut site, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRS
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
3JSX
 
 | | X-ray Crystal structure of NAD(P)H: Quinone Oxidoreductase-1 (NQO1) bound to the coumarin-based inhibitor AS1 | | Descriptor: | 4-hydroxy-6,7-dimethyl-3-(naphthalen-1-ylmethyl)-2H-chromen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Dunstan, M.S, Levy, C, Leys, D. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and biological evaluation of coumarin-based inhibitors of NAD(P)H: quinone oxidoreductase-1 (NQO1).
J.Med.Chem., 52, 2009
|
|
8DRT
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRW
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp9-nsp10 (C9) cut site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp9-nsp10 (C9) cut site, PENTAETHYLENE GLYCOL, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRR
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp4-nsp5 (C4) cut site sequence | | Descriptor: | 3C-like proteinase nsp5, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
2WOW
 
 | | Trypanosoma brucei trypanothione reductase with NADP and trypanothione bound | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-30 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WYO
 
 | | Trypanosoma brucei glutathione synthetase | | Descriptor: | GLUTATHIONE, GLUTATHIONE SYNTHETASE, SULFATE ION | | Authors: | Fyfe, P.K, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2009-11-17 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of Trypanosoma Brucei Glutathione Synthetase; Domain and Loop Alterations in the Catalytic Cycle of a Highly Conserved Enzyme.
Mol.Biochem.Parasitol., 170, 2010
|
|
2WP5
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00065414) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
8DRU
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp7-nsp8 (C7) cut site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp7-nsp8 (C7) cut site, PENTAETHYLENE GLYCOL, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8EBG
 
 | | Crystal structure of the probable FhuD FeIII-dicitrate-binding domain protein FecB from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, FEIII-dicitrate-binding periplasmic lipoprotein FecB, FORMIC ACID, ... | | Authors: | Cuff, M, Kim, Y, Endres, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2022-08-31 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of the probable FhuD FeIII-dicitrate-binding domain protein FecB from Mycobacterium tuberculosis
To Be Published
|
|
