7XIS
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | (2-methoxy-4-methoxycarbonyl-phenyl) 5-nitrofuran-2-carboxylate, MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XJ4
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, S-[5-[(E)-2-phenylethenyl]-1,3,4-oxadiazol-2-yl] 5-nitrothiophene-2-carbothioate, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7WU2
 
 | | Cryo-EM structure of the adhesion GPCR ADGRD1 in complex with miniGs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU4
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGi | | Descriptor: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
4WOI
 
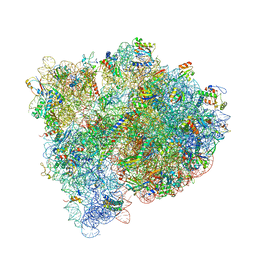 | | 4,5-linked aminoglycoside antibiotics regulate the bacterial ribosome by targeting dynamic conformational processes within intersubunit bridge B2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pulk, A, Cate, J.H.D, Blanchard, S, Wasserman, M, Altman, R, Zhou, Z, Zinder, J, Green, K, Garneau-Tsodikova, S. | | Deposit date: | 2014-10-15 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemically related 4,5-linked aminoglycoside antibiotics drive subunit rotation in opposite directions.
Nat Commun, 6, 2015
|
|
3CSC
 
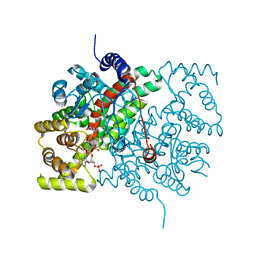 | |
3CUJ
 
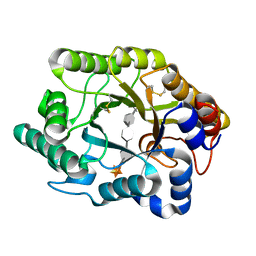 | |
7XNG
 
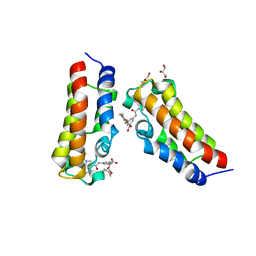 | | Crystal structure of CBP bromodomain liganded with Y08092(31g) | | Descriptor: | 3-[(1-ethanoylindol-3-yl)carbonylamino]-5-[[(2S)-oxan-2-yl]oxymethyl]benzoic acid, CREB-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08092(31g)
To Be Published
|
|
3CG0
 
 | | Crystal structure of signal receiver domain of modulated diguanylate cyclase from Desulfovibrio desulfuricans G20, an example of alternate folding | | Descriptor: | Response regulator receiver modulated diguanylate cyclase with PAS/PAC sensor | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Chang, S, Groshong, C, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Modulated Diguanylate Cyclase from Desulfovibrio desulfuricans.
To be Published
|
|
3D4C
 
 | |
3D54
 
 | | Structure of PurLQS from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Formylglycinamide ribonucleotide amidotransferase, Phosphoribosylformylglycinamidine synthase 1, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2008-05-15 | | Release date: | 2008-07-22 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima: structural insights into complex formation.
Biochemistry, 47, 2008
|
|
3CJZ
 
 | | Effects of N2,N2-dimethylguanosine on RNA structure and stability: crystal structure of an RNA duplex with tandem m22G:A pairs | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*GP*(M2G)P*AP*CP*GP*UP*CP*CP*U)-3') | | Authors: | Pallan, P.S, Kreutz, C, Bosio, S, Micura, R, Egli, M. | | Deposit date: | 2008-03-14 | | Release date: | 2008-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effects of N2,N2-dimethylguanosine on RNA structure and stability: crystal structure of an RNA duplex with tandem m2 2G:A pairs.
Rna, 14, 2008
|
|
3CLV
 
 | | Crystal Structure of Rab5a from plasmodium falciparum, PFB0500c | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Rab5 protein, ... | | Authors: | Chattopadhyay, D, Wernimont, A.K, Langsley, G, Lew, J, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Sukumar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-20 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Rab5a from plasmodium falciparum, PFB0500c
To be Published
|
|
3CM1
 
 | |
3CEU
 
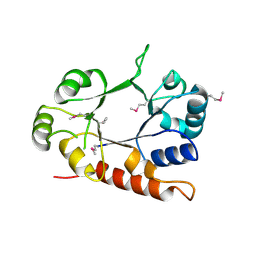 | | Crystal structure of thiamine phosphate pyrophosphorylase (BT_0647) from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium target BtR268 | | Descriptor: | Thiamine phosphate pyrophosphorylase | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Janjua, H, Mao, L, Xiao, R, Foote, E.L, Maglaqui, M, Ciccosanti, C, Zhao, L, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of thiamine phosphate pyrophosphorylase (BT_0647) from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium target BtR268.
To be Published
|
|
3CG6
 
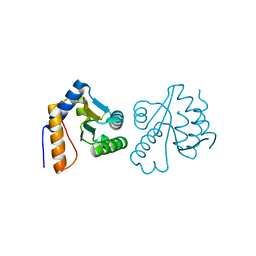 | | Crystal structure of Gadd45 gamma | | Descriptor: | Growth arrest and DNA-damage-inducible 45 gamma | | Authors: | Schrag, J.D, Jiralerspong, S, Banville, M, Jaramillo, M.L, O'Connor-McCourt, M.D. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure and dimerization interface of GADD45gamma.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7XM7
 
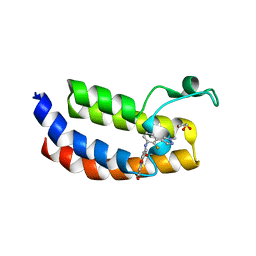 | | Crystal Structure of the CBP in complex with the Y08188 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethanoyl-~{N}-[2-fluoranyl-3-(1-methylpyrazol-4-yl)phenyl]-7-methoxy-indolizine-1-carboxamide, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as potent and selective CBP bromodomain inhibitors
To Be Published
|
|
3CJ7
 
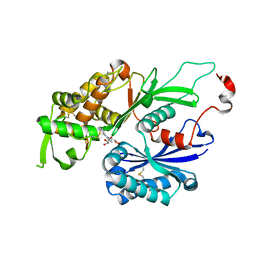 | | Structure of Rattus norvegicus NTPDase2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ectonucleoside triphosphate diphosphohydrolase 2 | | Authors: | Zebisch, M, Strater, N. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into signal conversion and inactivation by NTPDase2 in purinergic signaling
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7WUX
 
 | | Crystal structure of AziU3/U2 complexed with (5S,6S)-O7-sulfo DADH from Streptomyces sahachiroi | | Descriptor: | (2S,5S,6S)-2,6-bis(azanyl)-5-oxidanyl-7-sulfooxy-heptanoic acid, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, AziU2, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
3DLX
 
 | | Crystal structure of human 3-oxoacid CoA transferase 1 | | Descriptor: | GLYCEROL, Succinyl-CoA:3-ketoacid-coenzyme A transferase 1 | | Authors: | Kavanagh, K.L, Shafqat, N, Yue, W.W, Picaud, S, Murray, J.W, Maclean, E.M, von Delft, F, Roos, A.K, Arrowsmith, C.H, Wikstrom, M, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human 3-oxoacid CoA transferase 1.
To be Published
|
|
6MT1
 
 | |
3DCW
 
 | | Use of Carbonic Anhydrase II, IX Active-Site Mimic, for the Purpose of Screening Inhibitors for Possible Anti-Cancer Properties | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Genis, C, Sippel, K.H, Case, N, Govindasamy, L, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2008-06-04 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a carbonic anhydrase IX active-site mimic to screen inhibitors for possible anticancer properties
Biochemistry, 48, 2009
|
|
7XN7
 
 | | RNA polymerase II elongation complex containing Spt4/5, Elf1, Spt6, Spn1 and Paf1C | | Descriptor: | Chromatin elongation factor SPT5, Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
3DOB
 
 | | Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans. | | Descriptor: | BETA-MERCAPTOETHANOL, Heat shock 70 kDa protein F44E5.5 | | Authors: | Osipiuk, J, Hatzos, C, Gu, M, Zhang, R, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-03 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray crystal structure of Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans.
To be Published
|
|
7WUW
 
 | | Crystal structure of AziU3/U2 from Streptomyces sahachiroi | | Descriptor: | AziU2, AziU3, MAGNESIUM ION, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
