8ZDX
 
 | | Crystal structure of MjHKU4r-CoV-1 RBD bound to hDPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yang, M, Li, Z, Xu, Y, Zhang, S. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of MjHKU4r-CoV-1 RBD bound to hDPP4
To Be Published
|
|
8XNO
 
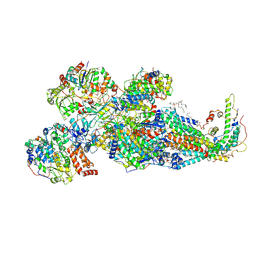 | | Respiratory complex Peripheral Arm of CI, close form A, focus-refined map of type IA, Wild type mouse under Cold Acclimation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 2024
|
|
4LOP
 
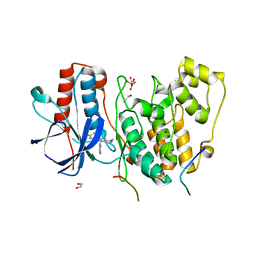 | | Structural basis of autoactivation of p38 alpha induced by TAB1 (Tetragonal crystal form) | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, L(+)-TARTARIC ACID, ... | | Authors: | Chaikuad, A, DeNicola, G.F, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Marber, M.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism and consequence of the autoactivation of p38 alpha mitogen-activated protein kinase promoted by TAB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
9EQJ
 
 | | Crystal structure of pVHL:EloB:EloC in complex with MP-1-39 | | Descriptor: | (2S,4R)-1-[(2R)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-3-[[cis-4-(morpholin-4-ylmethyl)cyclohexyl]methylsulfanyl]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kroupova, A, Pierri, M, Liu, X, Ciulli, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Stereochemical inversion at a 1,4-cyclohexyl PROTAC linker fine-tunes conformation and binding affinity.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
9CER
 
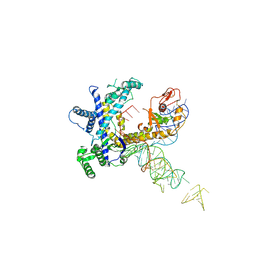 | | Guillardia theta Fanzor (GtFz) State 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, RNA (142-MER), ZINC ION | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
4LPF
 
 | | Crystal structure of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Ahangar, M.S, Vyas, R, Nasir, N, Biswal, B.K. | | Deposit date: | 2013-07-16 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the native, substrate-
bound and inhibited forms of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase
Acta Crystallogr.,Sect.D, 2013
|
|
8XNS
 
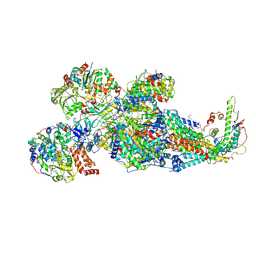 | | Respiratory complex Peripheral Arm of CI, close form B, focus-refined map of type IB, Wild type mouse under Cold Acclimation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 2024
|
|
9CET
 
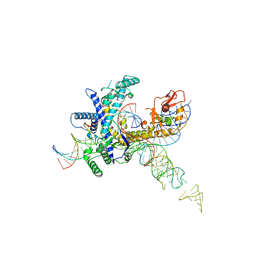 | | Guillardia theta Fanzor (GtFz) State 3 | | Descriptor: | DNA (28-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9FEF
 
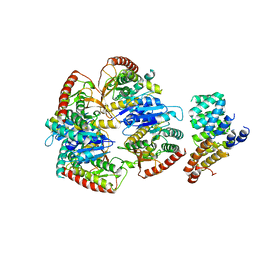 | | Cryo-EM structure of Trypanosoma cruzi (MDH)4-PEX5 complex | | Descriptor: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | Authors: | Lipinski, O, Sonani, R.R, Blat, A, Jemiola-Rzeminska, M, Patel, S.N, Sood, T, Dubin, G. | | Deposit date: | 2024-05-19 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM structure of Trypanosoma cruzi (MDH)4-PEX5 complex
To Be Published
|
|
9BZN
 
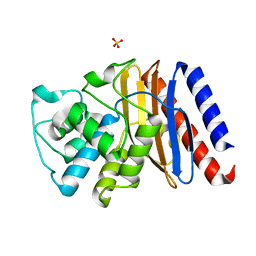 | | High resolution structure of class A Beta-lactamase from Bordetella bronchiseptica RB50 | | Descriptor: | FORMIC ACID, SULFATE ION, class A Beta-lactamase, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-05-24 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | High resolution structure of class A Beta-lactamase from Bordetella bronchiseptica RB50
To Be Published
|
|
9FX6
 
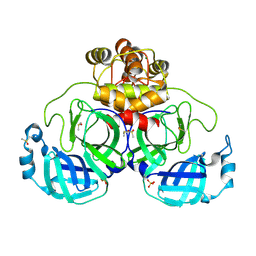 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 100K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9GNL
 
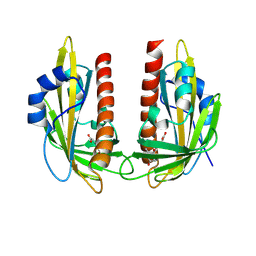 | | X-ray crystal structure of VvPYL1-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Rivera-Moreno, M, Benavente, J.L, Infantes, L, Albert, A. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of transpiration in grapevine through chemical activation of ABA signaling by ABA receptor agonists
To Be Published
|
|
8ZMF
 
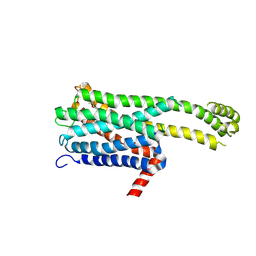 | | Crystal structure of an inverse agonist antipsychotic drug derivative-bound 5-HT2C | | Descriptor: | 1-[(4-fluorophenyl)methyl]-1-[(8~{S})-5-methyl-5-azaspiro[2.5]octan-8-yl]-3-[[4-(2-methylpropoxy)phenyl]methyl]urea, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562 | | Authors: | Oguma, T, Asada, H, Sekiguchi, Y, Imono, M, Iwata, S, Kusakabe, K. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Dual 5-HT 2A and 5-HT 2C Receptor Inverse Agonist That Affords In Vivo Antipsychotic Efficacy with Minimal hERG Inhibition for the Treatment of Dementia-Related Psychosis.
J.Med.Chem., 67, 2024
|
|
9AVJ
 
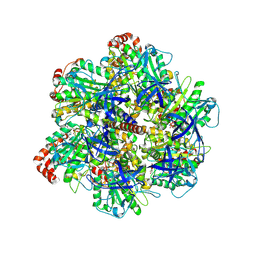 | | PS3 F1 ATPase Wild type | | Descriptor: | ATP synthase gamma chain, ATP synthase subunit alpha, ATP synthase subunit beta, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2024-03-03 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | The molecular structure of an axle-less F 1 -ATPase.
Biochim Biophys Acta Bioenerg, 2024
|
|
9CEV
 
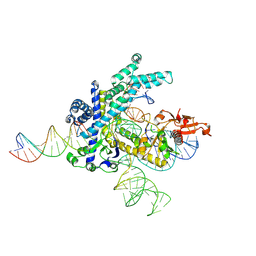 | | Spizellomyces punctatus Fanzor (SpuFz) State 2 | | Descriptor: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9G5L
 
 | |
9CEY
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 5 | | Descriptor: | DNA (26-MER), DNA (36-MER), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9DQC
 
 | |
9ERX
 
 | | Structural basis of D9-THC analog activity at the Cannabinoid 1 receptor | | Descriptor: | (6aR,10aR)-9-(hydroxymethyl)-6,6-dimethyl-3-(2-methyloctan-2-yl)-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, Antibody ScFv16 Fab fragment, Cannabinoid receptor 1, ... | | Authors: | Thorsen, T.S, Kulkarni, Y, Boggild, A, Drace, T, Nissen, P, Gajhede, M, Boesen, T, Kastrup, J.S, Gloriam, D. | | Deposit date: | 2024-03-25 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of Delta 9 -THC analog activity at the Cannabinoid 1 receptor.
Res Sq, 2024
|
|
9FCY
 
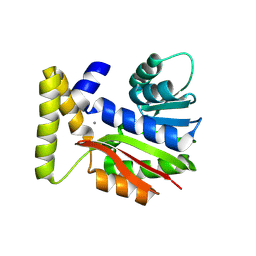 | | CysG(N-16)-R21K mutant in complex with SAH from Kitasatospora cystarginea | | Descriptor: | ADENINE, CALCIUM ION, SAM-dependent methyltransferase | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-16 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
9FX7
 
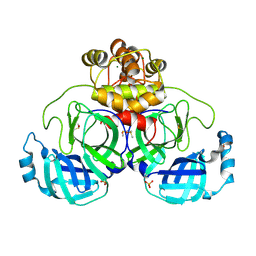 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 294K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9G3X
 
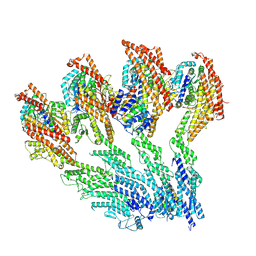 | |
8ZYP
 
 | | Cryo-EM Structure of E-4031-bound hERG Channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 2, ~{N}-[4-[1-[2-(6-methylpyridin-2-yl)ethyl]piperidin-4-yl]carbonylphenyl]methanesulfonamide | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
8XNQ
 
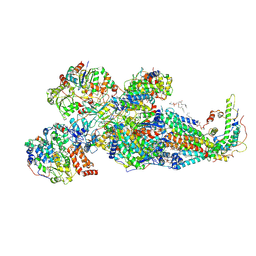 | | Respiratory complex Peripheral Arm of CI, open form A, focus-refined map of type IA, Wild type mouse under Cold Acclimation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 2024
|
|
8XNM
 
 | | Respiratory complex Peripheral Arm of CI, close form B, focus-refined map of type I, Wild type mouse under thermoneutral temperature | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 2024
|
|
