6ZJP
 
 | |
2R29
 
 | | Neutralization of dengue virus by a serotype cross-reactive antibody elucidated by cryoelectron microscopy and x-ray crystallography | | Descriptor: | Envelope protein E, Heavy chain of Fab 1A1D-2, Light chain of Fab 1A1D-2 | | Authors: | Lok, S.M, Kostyuchenko, V.K, Nybakken, G.E, Holdaway, H.A, Battisti, A.J, Sukupolvi-petty, S, Sedlak, D, Fremont, D.H, Chipman, P.R, Roehrig, J.T, Diamond, M.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-08-24 | | Release date: | 2007-12-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6AWU
 
 | | Structure of PR 10 Allergen Ara h 8.01 in complex with caffeic acid | | Descriptor: | Ara h 8 allergen, CAFFEIC ACID, CHLORIDE ION, ... | | Authors: | Offermann, L.R, McBride, J, Perdue, M, Hurlburt, B.K, Maleki, S.J, Pote, S.S, Chruszcz, M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of PR-10 Allergen Ara h 8.01.
To Be Published
|
|
6N88
 
 | |
8QJE
 
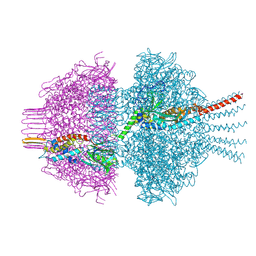 | | Neck of phage 812 virion (C12) | | Descriptor: | Portal protein, Putative neck protein, ZINC ION | | Authors: | Cienikova, Z, Novacek, J, Fuzik, T, Benesik, M, Plevka, P. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Genome anchoring, retention, and release by neck proteins of Herelleviridae phage 812
To Be Published
|
|
3EV0
 
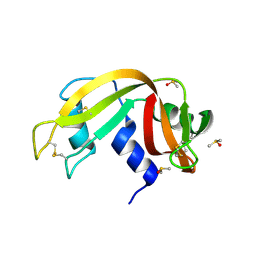 | | Crystal Structure of Ribonuclease A in 70% Dimethyl Sulfoxide | | Descriptor: | DIMETHYL SULFOXIDE, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
6ZJR
 
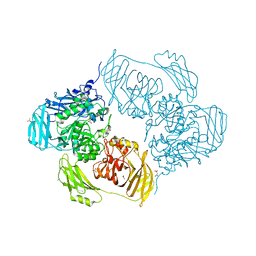 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with lactulose | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
2R1Z
 
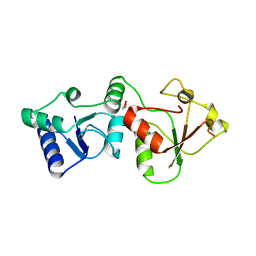 | |
6ZJS
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with galactose | | Descriptor: | (2S)-2-hydroxybutanedioic acid, ACETATE ION, Beta-galactosidase, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
8QKH
 
 | | Neck of phage 812 virion (C6) | | Descriptor: | Baseplate hub assembly protein, Capsid protein, Non-cytoplasmic protein, ... | | Authors: | Cienikova, Z, Novacek, J, Fuzik, T, Benesik, M, Plevka, P. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Genome anchoring, retention, and release by neck proteins of Herelleviridae phage 812
To Be Published
|
|
3EAM
 
 | | An open-pore structure of a bacterial pentameric ligand-gated ion channel | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Bocquet, N, Nury, H, Baaden, M, Le Poupon, C, Changeux, J.P, Delarue, M, Corringer, P.J. | | Deposit date: | 2008-08-26 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation.
Nature, 457, 2009
|
|
8A82
 
 | | Fe(II)/aKG-dependent halogenase OocPQ | | Descriptor: | Cupin_8 domain-containing protein, FE (III) ION, GLYCEROL, ... | | Authors: | Fraley, A.E, Meoded, R.A, Schmalhofer, M, Bergande, C, Groll, M, Piel, J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Heterocomplex structure of a polyketide synthase component involved in modular backbone halogenation.
Structure, 31, 2023
|
|
6YXV
 
 | |
6YVY
 
 | |
6YVS
 
 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH 5-{4-[(Pyridin-3-ylmethyl)-amino]-5-trifluoromethyl-pyrimidin-2-ylamino}-1,3-dihydro-indol-2-one | | Descriptor: | 5-[[4-(pyridin-3-ylmethylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino]-1,3-dihydroindol-2-one, Focal adhesion kinase 1, SULFATE ION | | Authors: | Musil, D, Heinrich, T, Amaral, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-kinetic relationship reveals the mechanism of selectivity of FAK inhibitors over PYK2.
Cell Chem Biol, 28, 2021
|
|
5M4L
 
 | | Crystal Structure of Wild-Type Human Prolidase with Mg ions and LeuPro ligand | | Descriptor: | GLYCEROL, HYDROXIDE ION, LEUCINE, ... | | Authors: | Wilk, P, Weiss, M.S, Mueller, U, Dobbek, H. | | Deposit date: | 2016-10-18 | | Release date: | 2017-07-12 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Substrate specificity and reaction mechanism of human prolidase.
FEBS J., 284, 2017
|
|
6AWV
 
 | | Ara h 8.01 in complex with epicatechin | | Descriptor: | (2R,3R)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol, Ara h 8 allergen, BENZOIC ACID, ... | | Authors: | Offermann, L.R, Perdue, M, McBride, J, Hurlburt, B.K, Maleki, S.J, Pote, S.S, Chruszcz, M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of PR-10 Allergen Ara h 8.01.
To Be Published
|
|
6YSY
 
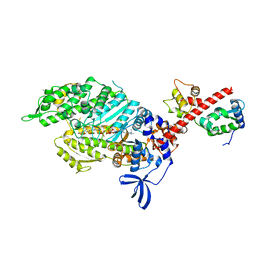 | | Skeletal Myosin bound to MPH-220, MgADP-VO4 | | Descriptor: | (9~{S})-5-methyl-12-(4-morpholin-4-ylphenyl)-9-oxidanyl-4-thia-2,12-diazatricyclo[7.3.0.0^{3,7}]dodeca-1,3(7),5-trien-8-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Canon, L, Kikuti, C.M, Gyimesi, M, Malnasi-Csizmadia, A, Houdusse, A. | | Deposit date: | 2020-04-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.246 Å) | | Cite: | Single Residue Variation in Skeletal Muscle Myosin Enables Direct and Selective Drug Targeting for Spasticity and Muscle Stiffness.
Cell, 183, 2020
|
|
5LOA
 
 | |
6D7J
 
 | |
8QX6
 
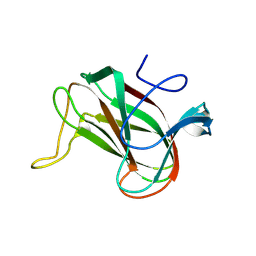 | | Novel laminarin-binding CBM X584 | | Descriptor: | PKD domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Zuehlke, M.K, Jeudy, A, Czjzek, M. | | Deposit date: | 2023-10-22 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ.Microbiol., 26, 2024
|
|
5LTM
 
 | |
6M5D
 
 | | Human serum albumin (apo form) | | Descriptor: | PHOSPHATE ION, Serum albumin | | Authors: | Ito, S, Senoo, A, Nagatoishi, S, Yamamoto, M, Tsumoto, K, Wakui, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Binding Mechanism of Human Serum Albumin Complexed with Cyclic Peptide Dalbavancin.
J.Med.Chem., 63, 2020
|
|
6LM1
 
 | | The crystal structure of cyanorhodopsin (CyR) N4075R from cyanobacteria Tolypothrix sp. NIES-4075 | | Descriptor: | DECANE, DODECANE, HEXADECANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2019-12-24 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique clade of light-driven proton-pumping rhodopsins evolved in the cyanobacterial lineage.
Sci Rep, 10, 2020
|
|
3CXG
 
 | | Crystal structure of Plasmodium falciparum thioredoxin, PFI0790w | | Descriptor: | GLYCEROL, Putative thioredoxin, SULFATE ION | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wilkstrom, M, Edwards, A.M, Hui, R, Hills, T, Pizarro, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-24 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Plasmodium falciparum thioredoxin, PFI0790w.
To be Published
|
|
