5PBU
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 15) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4X68
 
 | | Crystal Structure of OP0595 complexed with AmpC | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, NICKEL (II) ION | | Authors: | Yamada, M, Watanabe, T. | | Deposit date: | 2014-12-07 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
4X9E
 
 | | DEOXYGUANOSINETRIPHOSPHATE TRIPHOSPHOHYDROLASE from Escherichia coli with two DNA effector molecules | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, MAGNESIUM ION, RNA (5'-R(P*CP*CP*C)-3') | | Authors: | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | Deposit date: | 2014-12-11 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
4WEM
 
 | | Co-complex structure of the F4 fimbrial adhesin FaeG variant ac with llama single domain antibody V1 | | Descriptor: | Anti-F4+ETEC bacteria VHH variable region, K88 fimbrial protein AC, PHOSPHATE ION | | Authors: | Moonens, K, Van den Broeck, I, Pardon, E, De Kerpel, M, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight in the inhibition of adherence of F4 fimbriae producing enterotoxigenic Escherichia coli by llama single domain antibodies.
Vet. Res., 46, 2015
|
|
3LI6
 
 | | Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica | | Descriptor: | CALCIUM ION, Calcium-binding protein | | Authors: | Kumar, S, Ahmad, E, Kumar, S, Mansuri, M.S, Khan, R.H, Samudrala, G. | | Deposit date: | 2010-01-24 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica
Biophys.J., 98, 2010
|
|
4WF9
 
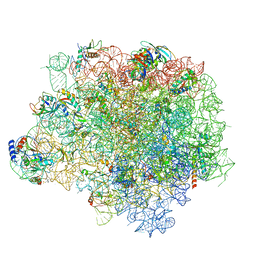 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with telithromycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.427 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WFB
 
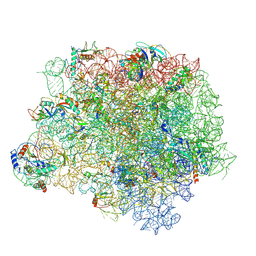 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with BC-3205 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WMG
 
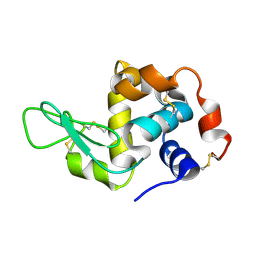 | | Structure of hen egg-white lysozyme from a microfludic harvesting device using synchrotron radiation (2.5A) | | Descriptor: | Lysozyme C | | Authors: | Lyubimov, A.Y, Murray, T.D, Koehl, A, Uervirojnangkoorn, M, Zeldin, O.B, Cohen, A.E, Soltis, S.M, Baxter, E.M, Brewster, A.S, Sauter, N.K, Brunger, A.T, Berger, J.M. | | Deposit date: | 2014-10-08 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capture and X-ray diffraction studies of protein microcrystals in a microfluidic trap array.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5YAS
 
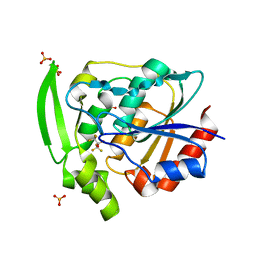 | | HYDROXYNITRILE LYASE COMPLEXED WITH HEXAFLUOROACETONE | | Descriptor: | 1,1,1,3,3,3-HEXAFLUOROPROPANEDIOL, PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
4X09
 
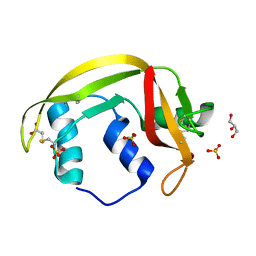 | | Structure of human RNase 6 in complex with sulphate anions | | Descriptor: | GLYCEROL, Ribonuclease K6, SULFATE ION | | Authors: | Prats-Ejarque, G, Arranz-Trullen, J, Blanco, J.A, Pulido, D, Moussaoui, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement.
Biochem.J., 473, 2016
|
|
4X0J
 
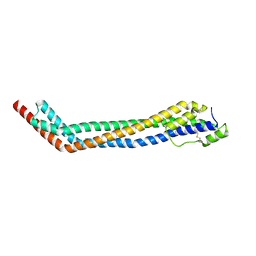 | | Trypanosoma brucei haptoglobin-haemoglobin receptor | | Descriptor: | Haptoglobin-hemoglobin receptor | | Authors: | Lane-Serff, H, MacGregor, P, Lowe, E.D, Carrington, M, Higgins, M.K. | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand and innate immunity factor uptake by the trypanosome haptoglobin-haemoglobin receptor.
Elife, 3, 2014
|
|
4X52
 
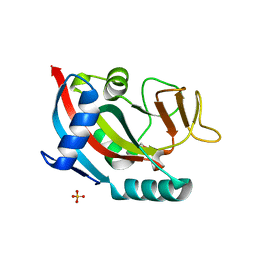 | | Human PARP13 (ZC3HAV1), C-Terminal PARP Domain (H810N; N830Y variant) | | Descriptor: | GLYCEROL, SULFATE ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Karlberg, T, Thorsell, A.G, Klepsch, M, Schuler, H. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
4WEU
 
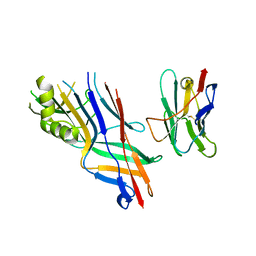 | | Co-complex structure of the F4 fimbrial adhesin FaeG variant ad with llama single domain antibody V3 | | Descriptor: | Anti-F4+ETEC bacteria VHH variable region, K88 fimbrial protein AD | | Authors: | Moonens, K, Van den Broeck, I, Pardon, E, De Kerpel, M, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insight in the inhibition of adherence of F4 fimbriae producing enterotoxigenic Escherichia coli by llama single domain antibodies.
Vet. Res., 46, 2015
|
|
4WGY
 
 | |
4WII
 
 | | HUMAN SPLICING FACTOR, CONSTRUCT 3 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Splicing factor, ... | | Authors: | lee, M, bond, c.s. | | Deposit date: | 2014-09-26 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation.
Nucleic Acids Res., 43, 2015
|
|
5YEG
 
 | | Crystal structure of CTCF ZFs4-8-Hs5-1a complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*AP*AP*CP*CP*AP*GP*CP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*GP*CP*TP*GP*GP*TP*TP*AP*AP*AP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Yin, M, Wang, J, Wang, M, Li, X. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
4WKZ
 
 | | Complex of autonomous ScaG cohesin CohG and X-doc domains | | Descriptor: | ACETATE ION, Autonomous cohesin, CALCIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Bayer, E.A, Yaniv, O, Shimon, L.J.W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Complex of autonomous ScaG cohesin CohG and X-doc domains
To Be Published
|
|
4WL7
 
 | | Room-temperature crystal structure of lysozyme determined by serial synchrotron crystallography using a micro focused beam (Conventional resolution cut-off) | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Coquelle, N, Brewster, A.S, Kapp, U, Shilova, A, Weinhausen, B, Burghammer, M, Colletier, J.P. | | Deposit date: | 2014-10-06 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WO0
 
 | | Crystal structure of transthyretin in complex with apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Transthyretin | | Authors: | Zanotti, G, Cianci, M, Folli, C, Berni, R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Structural evidence for asymmetric ligand binding to transthyretin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRI
 
 | | Crystal structure of okadaic acid binding protein 2.1 | | Descriptor: | OKADAIC ACID, Okadaic acid binding protein 2-alpha | | Authors: | Ehara, H, Makino, M, Kodama, K, Ito, T, Sekine, S, Fukuzawa, S, Yokoyama, S, Tachibana, K. | | Deposit date: | 2014-10-24 | | Release date: | 2015-05-27 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Okadaic Acid Binding Protein 2.1: A Sponge Protein Implicated in Cytotoxin Accumulation
Chembiochem, 16, 2015
|
|
4WSZ
 
 | |
4WT0
 
 | |
4WT3
 
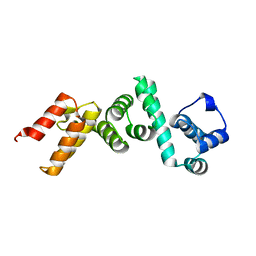 | | The N-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT4
 
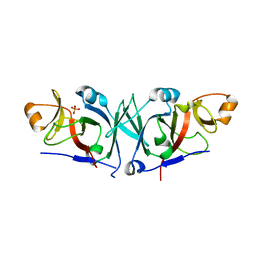 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form I | | Descriptor: | PHOSPHATE ION, Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT5
 
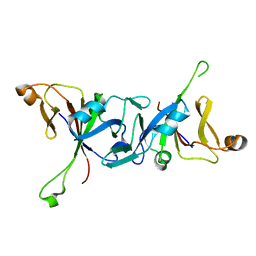 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form II | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
