7OII
 
 | | CspA-70 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
8XCS
 
 | | Cryo-EM structure of Glutamate dehydrogenase from Thermococcus profundus in complex with NADPH, AKG and NH4 in the initial stage of reaction | | Descriptor: | 2-OXOGLUTARIC ACID, AMMONIUM ION, Glutamate dehydrogenase, ... | | Authors: | Wakabayashi, T, Oide, M, Nakasako, M. | | Deposit date: | 2023-12-10 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | CryoEM-sampling of metastable conformations appearing in cofactor-ligand association and catalysis of glutamate dehydrogenase.
Sci Rep, 14, 2024
|
|
8XCY
 
 | |
8XD6
 
 | | Cryo-EM structure of Glutamate dehydrogenase from Thermococcus profundus in complex with NADPH, AKG and NH4 in the steady stage of reaction | | Descriptor: | 2-OXOGLUTARIC ACID, AMMONIUM ION, Glutamate dehydrogenase, ... | | Authors: | Wakabayashi, T, Oide, M, Nakasako, M. | | Deposit date: | 2023-12-10 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | CryoEM-sampling of metastable conformations appearing in cofactor-ligand association and catalysis of glutamate dehydrogenase.
Sci Rep, 14, 2024
|
|
8XD3
 
 | |
8XCP
 
 | |
8XCT
 
 | |
8XCZ
 
 | |
8X7T
 
 | | MCM in the Apo state. | | Descriptor: | mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | MCM in the Apo state
To Be Published
|
|
8X7U
 
 | | MCM in complex with dsDNA in presence of ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | MCM in complex with dsDNA in presence of ATP
To Be Published
|
|
8X36
 
 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form B) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8X35
 
 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8X37
 
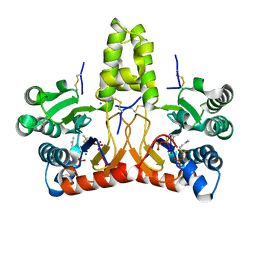 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8XJ0
 
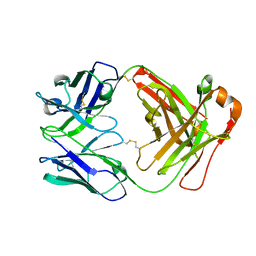 | | Crystal structure of AmFab mutant - P40C/E165C (Light chain), G10C/P210C(Heavy chain) | | Descriptor: | Adalimumab Fab Heavy chain, Adalimumab Fab Light chain | | Authors: | Senda, M, Yoshikawa, M, Nakamura, H, Ohkuri, T, Senda, T. | | Deposit date: | 2023-12-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Stabilization of adalimumab Fab through the introduction of disulfide bonds between the variable and constant domains.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
4UIR
 
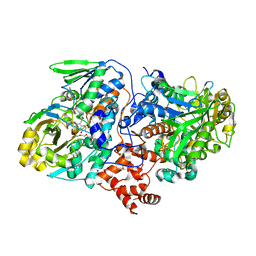 | | Structure of oleate hydratase from Elizabethkingia meningoseptica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HEXAETHYLENE GLYCOL, OLEATE HYDRATASE, ... | | Authors: | Pavkov-Keller, T, Hromic, A, Engleder, M, Emmerstorfer, A, Steinkellner, G, Schrempf, S, Wriessnegger, T, Leitner, E, Strohmeier, G.A, Kaluzna, I, Mink, D, Schuermann, M, Wallner, S, Macheroux, P, Pichler, H, Gruber, K. | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Mechanism of Oleate Hydratase from Elizabethkingia Meningoseptica.
Chembiochem, 16, 2015
|
|
8I5L
 
 | |
8WUC
 
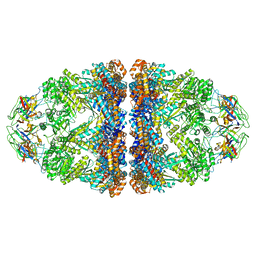 | | Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8UDU
 
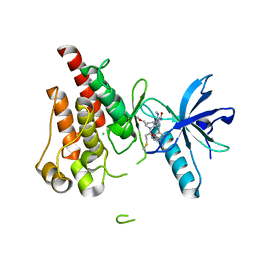 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8UDT
 
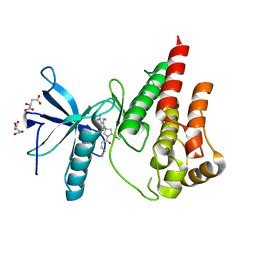 | | The X-RAY co-crystal structure of human FGFR3 and KIN-3248 | | Descriptor: | 3-[(1-cyclopropyl-4,6-difluoro-1H-benzimidazol-5-yl)ethynyl]-1-[(3R,5R)-5-(methoxymethyl)-1-propanoylpyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, D-MALATE, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.829 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
4UET
 
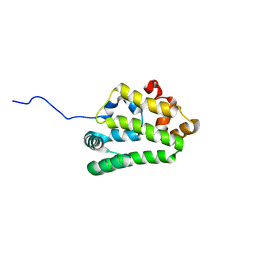 | | Diversity in the structures and ligand binding sites among the fatty acid and retinol binding proteins of nematodes revealed by Na-FAR-1 from Necator americanus | | Descriptor: | NEMATODE FATTY ACID RETINOID BINDING PROTEIN | | Authors: | Rey-Burusco, M.F, Ibanez Shimabukuro, M, Griffiths, K, Cooper, A, Kennedy, M.W, Corsico, B, Smith, B.O, Griffiths, K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Diversity in the Structures and Ligand Binding Sites of Nematode Fatty Acid and Retinol Binding Proteins Revealed by Na-Far-1 from Necator Americanus.
Biochem.J., 471, 2015
|
|
8WUW
 
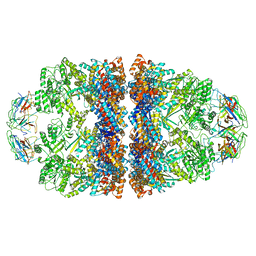 | | Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WUX
 
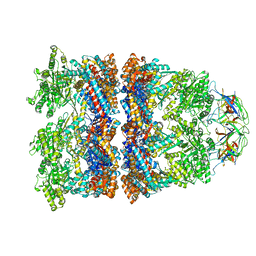 | | Cryo-EM structure of H. thermophilus GroEL-GroES bullet complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WU4
 
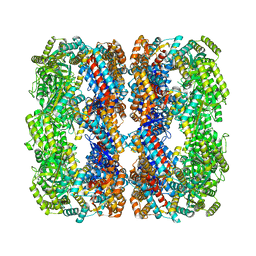 | | Cryo-EM structure of native H. thermoluteolus TH-1 GroEL | | Descriptor: | Chaperonin GroEL | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
