8I5F
 
 | | Crystal structure of the DHR-2 domain of DOCK10 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 10 | | Authors: | Kukimoto-Niino, M, Mishima-Tsumagari, C, Fukui, Y, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2023-01-25 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the dual GTPase specificity of the DOCK10 guanine nucleotide exchange factor.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
4WS2
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 6-aminouracil, Form I | | Descriptor: | 6-aminopyrimidine-2,4(3H,5H)-dione, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UPL
 
 | | Dimeric sulfatase SpAS2 from Silicibacter pomeroyi | | Descriptor: | SULFATASE FAMILY PROTEIN, ZINC ION | | Authors: | Jonas, S, van Loo, B, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Balancing Specificity and Promiscuity in Enzyme Evolution: Multidimensional Activity Transitions in the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 141, 2019
|
|
4WS7
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-chlorouracil, Form II | | Descriptor: | 1,2-ETHANEDIOL, 5-chloropyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8I5V
 
 | | DOCK10 mutant L1903Y complexed with Rac1 | | Descriptor: | Dedicator of cytokinesis protein 10, Ras-related C3 botulinum toxin substrate 1, SULFATE ION | | Authors: | Kukimoto-Niino, M, Mishima-Tsumagari, C, Ihara, K, Fukui, Y, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | Structural basis for the dual GTPase specificity of the DOCK10 guanine nucleotide exchange factor.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
4WPL
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with uracil, Form I | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UN2
 
 | | Crystal structure of the UBA domain of Dsk2 in complex with Ubiquitin | | Descriptor: | UBIQUITIN, UBIQUITIN DOMAIN-CONTAINING PROTEIN DSK2 | | Authors: | Michielssens, S, Peters, J.H, Ban, D, Pratihar, S, Seeliger, D, Sharma, M, Giller, K, Sabo, T.M, Becker, S, Lee, D, Griesinger, C, de Groot, B.L. | | Deposit date: | 2014-05-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A Designed Conformational Shift to Control Protein Binding Specificity.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UJ4
 
 | | Crystal structure of human Rab11-Rabin8-FIP3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rab-3A-interacting protein, ... | | Authors: | Vetter, M, Lorentzen, E. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of Rab11-FIP3-Rabin8 reveals simultaneous binding of FIP3 and Rabin8 effectors to Rab11.
Nat. Struct. Mol. Biol., 22, 2015
|
|
4UOB
 
 | | Crystal structure of Deinococcus radiodurans Endonuclease III-3 | | Descriptor: | ACETATE ION, COBALT (II) ION, ENDONUCLEASE III-3, ... | | Authors: | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | Deposit date: | 2014-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
4UN9
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 8H INCUBATION IN 5MM MN (STATE 3) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, MANGANESE (II) ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UJ3
 
 | | Crystal structure of human Rab11-Rabin8-FIP3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RAB-3A-INTERACTING PROTEIN, ... | | Authors: | Vetter, M, Lorentzen, E. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Rab11-Fip3-Rabin8 Reveals Simultaneous Binding of Fip3 and Rabin8 Effectors to Rab11.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UN5
 
 | | Crystal structure of Cas9 bound to PAM-containing DNA target containing mismatches at positions 1-3 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
4UN4
 
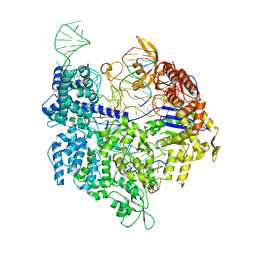 | | Crystal structure of Cas9 bound to PAM-containing DNA target with mismatches at positions 1-2 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.371 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
4UJ0
 
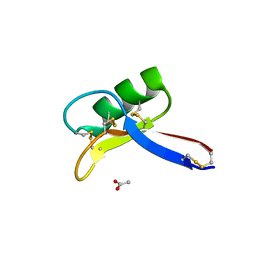 | | Crystal structure of the tomato defensin TPP3 | | Descriptor: | ACETATE ION, FLOWER-SPECIFIC GAMMA-THIONIN-LIKE PROTEIN/ACIDIC PROTEIN | | Authors: | Richter, V, Lay, F.T, Hulett, M.D, Kvansakul, M. | | Deposit date: | 2015-04-07 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Tomato Defensin Tpp3 Binds Phosphatidylinositol (4,5)-Bisphosphate Via a Conserved Dimeric Cationic Grip Conformation to Mediate Cell Lysis.
Mol.Cell.Biol., 35, 2015
|
|
4UMV
 
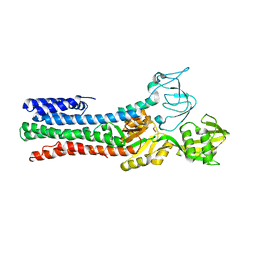 | | CRYSTAL STRUCTURE OF A ZINC-TRANSPORTING PIB-TYPE ATPASE IN THE E2P STATE | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ZINC-TRANSPORTING ATPASE | | Authors: | Wang, K.T, Sitsel, O, Meloni, G, Autzen, H.E, Andersson, M, Klymchuk, T, Nielsen, A.M, Rees, D.C, Nissen, P, Gourdon, P. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Mechanism of Zn(2+)-Transporting P-Type Atpases.
Nature, 514, 2014
|
|
4UOU
 
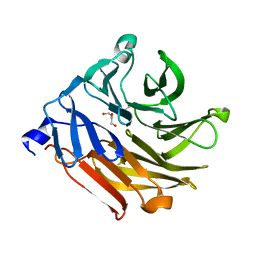 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) - apo-form | | Descriptor: | DI(HYDROXYETHYL)ETHER, FUCOSE-SPECIFIC LECTIN FLEA | | Authors: | Houser, J, Komarek, J, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2014-06-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5Y5M
 
 | | SFX structure of cytochrome P450nor: a complete dark data without pump laser (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
4UHM
 
 | | Characterization of a Novel Transaminase from Pseudomonas sp. Strain AAC | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Wilding, M, Peat, T.S, Newman, J, Scott, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A Beta-Alanine Catabolism Pathway Containing a Highly Promiscuous Omega-Transaminase in the 12-Aminododecanate-Degrading Pseudomonas Sp. Strain Aac.
Appl.Environ.Microbiol., 82, 2016
|
|
4UJ8
 
 | | Structure of surface layer protein SbsC, domains 6-7 | | Descriptor: | CALCIUM ION, SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Surface Layer Protein Sbsc
To be Published
|
|
1AX9
 
 | | ACETYLCHOLINESTERASE COMPLEXED WITH EDROPHONIUM, LAUE DATA | | Descriptor: | ACETYLCHOLINESTERASE, EDROPHONIUM ION | | Authors: | Raves, M.L, Ravelli, R.B.G, Sussman, J.L, Harel, M, Silman, I. | | Deposit date: | 1997-11-03 | | Release date: | 1998-02-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Static Laue diffraction studies on acetylcholinesterase.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
4UPI
 
 | | Dimeric sulfatase SpAS1 from Silicibacter pomeroyi | | Descriptor: | SULFATASE FAMILY PROTEIN, ZINC ION | | Authors: | Jonas, S, van Loo, B, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Balancing Specificity and Promiscuity in Enzyme Evolution: Multidimensional Activity Transitions in the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 141, 2019
|
|
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
4WJO
 
 | | Crystal Structure of SUMO1 in complex with PML | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
7AKI
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-NH2 | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxamide, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2020-10-01 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
