4XOX
 
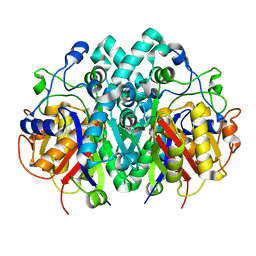 | | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae | | Descriptor: | 3-oxoacyl-ACP synthase | | Authors: | Hou, J, Grabowski, M, Cymborowski, M, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae
to be published
|
|
4XNI
 
 | | X-ray structure of PepTst1 | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UFZ
 
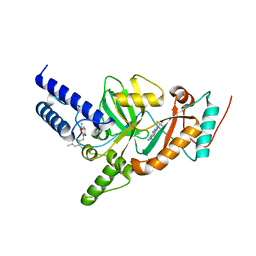 | | Synthesis of Novel NAD Dependant DNA Ligase Inhibitors via Negishi Cross-Coupling: Development of SAR and Resistance Studies | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5,7-bis(azanyl)-2-tert-butyl-4-(1,3-thiazol-2-yl)pyrido[2,3-d]pyrimidine-6-carbonitrile, DNA LIGASE | | Authors: | Murphy-Benenato, K.E, Boriack-Sjodin, P.A, Martinez-Botella, G, Carcanague, D, Gingipali, L, Gowravaram, M, Harang, J, Hale, M, Ioannidis, G, Jahic, H, Johnstone, M, Kutschke, A, Laganas, V.A, Loch, J, Oguto, H, Patel, S.J. | | Deposit date: | 2015-03-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Negishi Cross-Coupling Enabled Synthesis of Novel Nad(+)-Dependent DNA Ligase Inhibitors and Sar Development.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
4XNK
 
 | | X-ray structure of AlgE1 | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Alginate production protein AlgE, ... | | Authors: | Ma, P, Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XSK
 
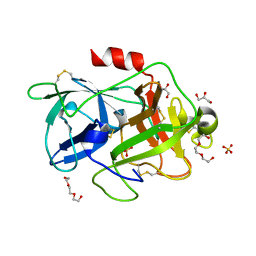 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
7AMF
 
 | |
7AMB
 
 | |
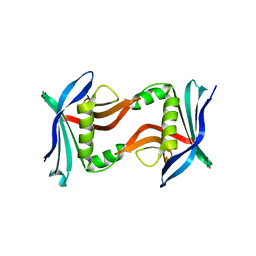
4U2I
 
 | |
7Q85
 
 | | Crystal structure of human STING in complex with MD1193 | | Descriptor: | 9-[(1R,6R,8R,13E,15R,17R,18R)-17-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,16-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadec-13-en-8-yl]purin-6-amine, Stimulator of interferon genes protein | | Authors: | Smola, M, Klima, M, Boura, E. | | Deposit date: | 2021-11-10 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Vinylphosphonate-based cyclic dinucleotides enhance STING-mediated cancer immunotherapy.
Eur.J.Med.Chem., 259, 2023
|
|
7QB6
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with 3-carboxybenzophenone | | Descriptor: | 3-benzoylbenzoic acid, MALONATE ION, Nodulin-13 | | Authors: | Grzechowiak, M, Ignasiak, M, Nowicka-Bauer, K, Marciniak, B, Jaskolski, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Does the presence of ground state complex between a PR-10 protein and a sensitizer affect the mechanism of sensitized photo-oxidation?
Free Radic Biol Med, 198, 2023
|
|
6XXR
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPPTEDEA-NH2 | | Descriptor: | Ac-[2-Cl-F]-PPPPTEDEA-NH2, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Le Cong, K, Roske, Y. | | Deposit date: | 2020-01-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8WOP
 
 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, wild-type | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase 2, URIDINE-5'-DIPHOSPHATE | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8RUF
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV D187A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
4U87
 
 | | Crystal structure of the Ba-soaked C2 crystal form of pMV158 replication initiator RepB (P3221 space group) | | Descriptor: | BARIUM ION, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Conformational plasticity of RepB, the replication initiator protein of promiscuous streptococcal plasmid pMV158.
Sci Rep, 6, 2016
|
|
8WOV
 
 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, G233A mutant | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase 2, URIDINE-5'-DIPHOSPHATE | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8WOW
 
 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, I160L/G233A mutant | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, UDP-glucose 4-epimerase 2, ... | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8RUD
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV K138A mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
7KGU
 
 | | Structure of 2Q1-Fab, an antibody selective for IDH2R140Q-HLA-B*07:02 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, M.S, Aytenfisu, T.Y, Wright, K.M, Gabelli, S.B. | | Deposit date: | 2020-10-18 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
6ZE8
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound OATD-01 | | Descriptor: | 5-(4-((2S,5S)-5-(4-chlorobenzyl)-2-methylmorpholino)piperidin-1-yl)-4H-1,2,4-triazol-3-amine, Chitotriosidase-1, GLYCEROL, ... | | Authors: | Nowotny, M, Bartlomiejczak, A, Napiorkowska-Gromadzka, A. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of OATD-01 , a First-in-Class Chitinase Inhibitor as Potential New Therapeutics for Idiopathic Pulmonary Fibrosis.
J.Med.Chem., 63, 2020
|
|
8JJS
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP10343 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAA-ILE-SAR-SAR-7T2-SAR-IAE-LEU-MEA-MLE-7TK, ... | | Authors: | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | Deposit date: | 2023-05-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Development of Orally Bioavailable Peptides Targeting an Intracellular Protein: From a Hit to a Clinical KRAS Inhibitor.
J.Am.Chem.Soc., 145, 2023
|
|
8IU0
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Nakamura, S, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2023-03-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
1B4T
 
 | | H48C YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 1999-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1B4L
 
 | | 15 ATMOSPHERE OXYGEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
