4QBV
 
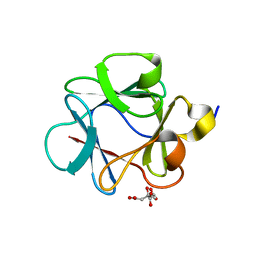 | |
4LH5
 
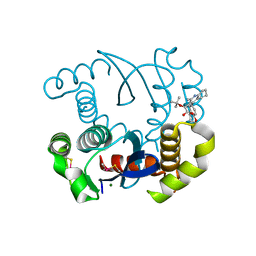 | | Dual inhibition of HIV-1 replication by Integrase-LEDGF allosteric inhibitors is predominant at post-integration stage during virus production rather than at integration | | Descriptor: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, Integrase, MAGNESIUM ION | | Authors: | Ruff, M, Levy, N, Eiler, S. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage.
Retrovirology, 10, 2013
|
|
2CYG
 
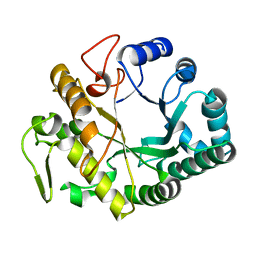 | | Crystal structure at 1.45- resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome | | Descriptor: | beta-1, 3-glucananse | | Authors: | Receveur-Brechot, V, Czjzek, M, Barre, A, Roussel, A, Peumans, W.J, Van Damme, E.J.M, Rouge, P. | | Deposit date: | 2005-07-07 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure at 1.45-A resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome
Proteins, 63, 2006
|
|
4Q3B
 
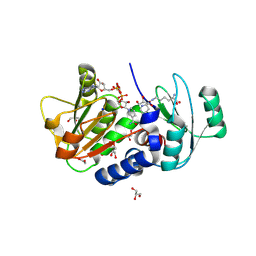 | | PylD cocrystallized with L-Lysine-Ne-D-lysine and NAD+ | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Quitterer, F, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2014-04-11 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Formation of Pyrroline and Tetrahydropyridine Rings in Amino Acids Catalyzed by Pyrrolysine Synthase (PylD).
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2CZ2
 
 | | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-1 crystal) | | Descriptor: | GLUTATHIONE, GLYCEROL, Maleylacetoacetate isomerase | | Authors: | Mizohata, E, Morita, S, Kinoshita, Y, Nagano, K, Uda, H, Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-10 | | Release date: | 2006-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-1 crystal)
To be Published
|
|
4LFE
 
 | | Crystal structure of geranylgeranyl diphosphate synthase sub1274 (target efi-509455) from streptococcus uberis 0140j with bound magnesium and isopentyl diphosphate, partially liganded complex; | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl diphosphate synthase, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Geranylgeranyl Diphosphate Synthase from Streptococcus Uberis 0140J
To be Published
|
|
2CZD
 
 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution
To be Published
|
|
1TIW
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-Tetrahydro-2-furoic acid | | Descriptor: | Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE, TETRAHYDROFURAN-2-CARBOXYLIC ACID | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-02 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1TJ0
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) co-crystallized with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-02 | | Release date: | 2004-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1TJK
 
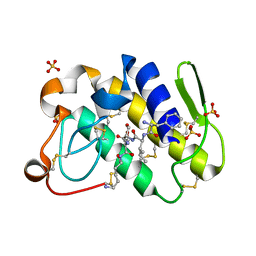 | | Crystal structure of the complex formed between group II phospholipase A2 with a designed pentapeptide, Phe- Leu- Ser- Thr- Lys at 1.2 A resolution | | Descriptor: | Phospholipase A2, SULFATE ION, synthetic peptide | | Authors: | Singh, N, Jabeen, T, Somvanshi, R.K, Sharma, S, Perbandt, M, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2004-06-06 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the complex formed between group II phospholipase A2 with a designed pentapeptide, Phe - Leu - Ser - Thr - Lys at 1.2 A resolution
To be Published
|
|
1TK7
 
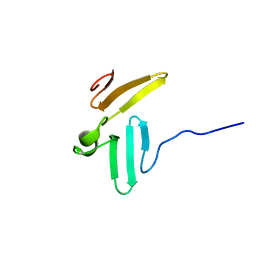 | | NMR structure of WW domains (WW3-4) from Suppressor of Deltex | | Descriptor: | CG4244-PB | | Authors: | Fedoroff, O.Y, Avis, J.M, Golovanov, A.P, Baron, M, Townson, S.A. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure and dynamics of tandem WW domains in a negative regulator of notch signaling, Suppressor of deltex
J.Biol.Chem., 279, 2004
|
|
1GNF
 
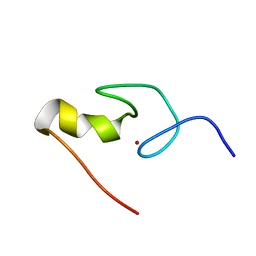 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZINC FINGER OF MURINE GATA-1, NMR, 25 STRUCTURES | | Descriptor: | TRANSCRIPTION FACTOR GATA-1, ZINC ION | | Authors: | Kowalski, K, Czolij, R, King, G.F, Crossley, M, Mackay, J.P. | | Deposit date: | 1998-10-12 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal zinc finger of GATA-1 reveals a specific binding face for the transcriptional co-factor FOG.
J.Biomol.NMR, 13, 1999
|
|
1TKU
 
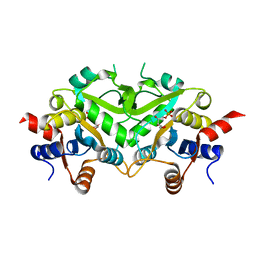 | | Crystal Structure of 3,4-Dihydroxy-2-butanone 4-phosphate Synthase of Candida albicans in complex with Ribulose-5-phosphate | | Descriptor: | 3,4-Dihydroxy-2-butanone 4-phosphate Synthase, RIBULOSE-5-PHOSPHATE | | Authors: | Echt, S, Bauer, S, Steinbacher, S, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2004-06-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone 4-phosphate synthase of Candida albicans.
J.Mol.Biol., 341, 2004
|
|
2CYZ
 
 | | photo-activation state of Fe-type NHase in anaerobic condition | | Descriptor: | FE (III) ION, MAGNESIUM ION, Nitrile hydratase subunit alpha, ... | | Authors: | Kawano, Y, Hashimoto, K, Odaka, M, Nakayama, H, Takio, K, Endo, I, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-09 | | Release date: | 2006-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | photo-activation state of Fe-type NHase in anaerobic condition
To be Published
|
|
1GGZ
 
 | | CRYSTAL STRUCTURE OF THE CALMODULIN-LIKE PROTEIN (HCLP) FROM HUMAN EPITHELIAL CELLS | | Descriptor: | CALCIUM ION, CALMODULIN-RELATED PROTEIN NB-1 | | Authors: | Han, B.-G, Han, M, Sui, H, Yaswen, P, Walian, P.J, Jap, B.K. | | Deposit date: | 2000-10-13 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human calmodulin-like protein: insights into its functional role.
FEBS Lett., 521, 2002
|
|
1GIH
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE CDK4 INHIBITOR | | Descriptor: | 1-(5-OXO-2,3,5,9B-TETRAHYDRO-1H-PYRROLO[2,1-A]ISOINDOL-9-YL)-3-PYRIDIN-2-YL-UREA, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Ikuta, M, Nishimura, S. | | Deposit date: | 2001-02-06 | | Release date: | 2002-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein.
J.Biol.Chem., 276, 2001
|
|
2CZF
 
 | |
4Q5E
 
 | |
2CLB
 
 | | The structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like di-metal binding site within a Dps- like dodecameric assembly | | Descriptor: | DPS-LIKE PROTEIN, FE (III) ION, ZINC ION | | Authors: | Gauss, G.H, Benas, P, Wiedenheft, B, Young, M, Douglas, T, Lawrence, C.M. | | Deposit date: | 2006-04-26 | | Release date: | 2006-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Dps-Like Protein from Sulfolobus Solfataricus Reveals a Bacterioferritin-Like Dimetal Binding Site within a Dps-Like Dodecameric Assembly.
Biochemistry, 45, 2006
|
|
4QFZ
 
 | | Crystal structure of the tetrameric dGTP/dTTP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
2D07
 
 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | Descriptor: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-07-26 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
1BCG
 
 | | SCORPION TOXIN BJXTR-IT | | Descriptor: | TOXIN BJXTR-IT | | Authors: | Oren, D, Froy, O, Amit, E, Kleinberger-Doron, N, Gurevitz, M, Shaanan, B. | | Deposit date: | 1998-04-29 | | Release date: | 1998-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An excitatory scorpion toxin with a distinctive feature: an additional alpha helix at the C terminus and its implications for interaction with insect sodium channels.
Structure, 6, 1998
|
|
4Q5W
 
 | | Crystal structure of extended-Tudor 9 of Drosophila melanogaster | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Maternal protein tudor | | Authors: | Ren, R, Liu, H, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
1ST8
 
 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Verhaest, M, Van den Ende, W, De Ranter, C.J, Van Laere, A, Rabijns, A. | | Deposit date: | 2004-03-25 | | Release date: | 2005-03-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus.
Plant J., 41, 2005
|
|
1GIQ
 
 | | Crystal Structure of the Enzymatic Componet of Iota-Toxin from Clostridium Perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, IOTA TOXIN COMPONENT IA | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
