3LYS
 
 | | Crystal Structure of the N-terminal domain of the Prophage pi2 protein 01 (integrase) from Lactococcus lactis, Northeast Structural Genomics Consortium Target KR124F | | Descriptor: | Prophage pi2 protein 01, integrase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-28 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Northeast Structural Genomics Consortium Target KR124F
To be Published
|
|
5CBW
 
 | | Human Cyclophilin D Complexed with Inhibitor. | | Descriptor: | FORMIC ACID, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor.
To Be Published
|
|
4X3F
 
 | |
2WCN
 
 | | Solution structure of an LNA-modified quadruplex | | Descriptor: | DNA (5'-D(*DGP*LCG*DGP*LCG*DTP*DTP*DTP *DTP*DGP*LCG*DGP*LCG)-3') | | Authors: | Nielsen, J.T, Arar, K, Petersen, M. | | Deposit date: | 2009-03-12 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Locked Nucleic Acid Modified Quadruplex: Introducing the V4 Folding Topology.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3LYU
 
 | | Crystal Structure of the C-terminal domain (residues 83-215) of PF1911 hydrogenase from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR246A | | Descriptor: | Putative hydrogenase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Foote, E.L, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-28 | | Release date: | 2010-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target PfR246A
To be Published
|
|
2W1Y
 
 | | THE INTERDEPENDENCE OF WAVELENGTH, REDUNDANCY AND DOSE IN SULFUR SAD EXPERIMENTS: 1.540 A wavelength 180 images data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1YBU
 
 | | Mycobacterium tuberculosis adenylyl cyclase Rv1900c CHD, in complex with a substrate analog. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION, lipJ | | Authors: | Sinha, S.C, Wetterer, M, Sprang, S.R, Schultz, J.E, Linder, J.U. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Origin of asymmetry in adenylyl cyclases: structures of Mycobacterium tuberculosis Rv1900c.
Embo J., 24, 2005
|
|
5CG6
 
 | | Neutron crystal structure of human farnesyl pyrophosphate synthase in complex with risedronate and isopentenyl pyrophosphate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Yokoyama, T, Mizuguchi, M, Ostermann, A, Kusaka, K, Niimura, N, Schrader, T.E, Tanaka, I. | | Deposit date: | 2015-07-09 | | Release date: | 2015-10-14 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Protonation State and Hydration of Bisphosphonate Bound to Farnesyl Pyrophosphate Synthase
J.Med.Chem., 58, 2015
|
|
4G26
 
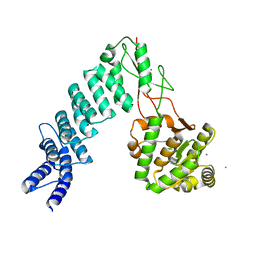 | | Crystal Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana with Ca | | Descriptor: | CALCIUM ION, Pentatricopeptide repeat-containing protein At2g32230, mitochondrial, ... | | Authors: | Koutmos, M, Howard, M.J, Fierke, C.A. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5' processing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1YCJ
 
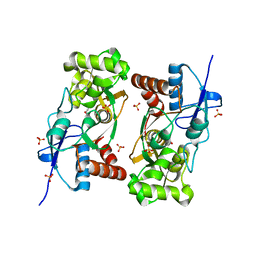 | | Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate | | Descriptor: | GLUTAMIC ACID, Ionotropic glutamate receptor 5, SULFATE ION | | Authors: | Naur, P, Vestergaard, B, Skov, L.K, Egebjerg, J, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2004-12-22 | | Release date: | 2005-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate
Febs Lett., 579, 2005
|
|
2W1L
 
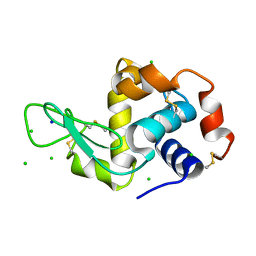 | | THE INTERDEPENDENCE OF WAVELENGTH, REDUNDANCY AND DOSE IN SULFUR SAD EXPERIMENTS: 0.979 a wavelength 991 images data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-17 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1Y4K
 
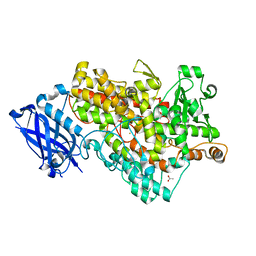 | | Lipoxygenase-1 (Soybean) at 100K, N694G Mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (II) ION, ... | | Authors: | Chruszcz, M, Segraves, E, Holman, T.R, Minor, W. | | Deposit date: | 2004-12-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Kinetic, spectroscopic, and structural investigations of the soybean lipoxygenase-1 first-coordination sphere mutant, Asn694Gly.
Biochemistry, 45, 2006
|
|
4G4J
 
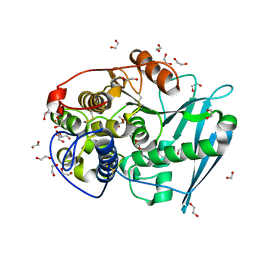 | | Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile in complex with methyl 4-O-methyl-beta-D-glucopyranuronate determined at 2.35 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-glucuronoyl methylesterase, GLYCEROL, ... | | Authors: | Charavgi, M.D, Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2012-07-16 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2VO6
 
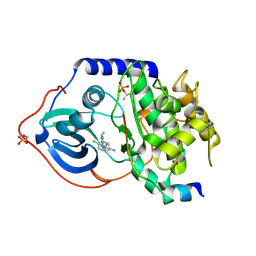 | | Structure of PKA-PKB chimera complexed with 4-(4-Chlorobenzyl)-1-(7H- pyrrolo(2,3-d)pyrimidin-4-yl)piperidin-4-ylamine | | Descriptor: | 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
5CJT
 
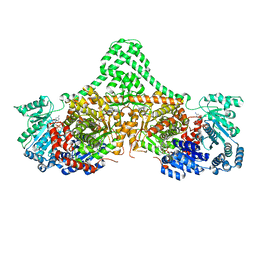 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, Mg (holo-IcmF/GDP), and substrate isobutyryl-coenzyme A | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for Substrate Specificity in Adenosylcobalamin-dependent Isobutyryl-CoA Mutase and Related Acyl-CoA Mutases.
J.Biol.Chem., 290, 2015
|
|
2VO3
 
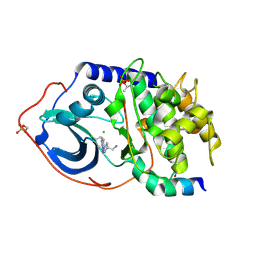 | | Structure of PKA-PKB chimera complexed with C-(4-(4-Chlorophenyl)-1-(7H-pyrrolo(2,3-d)pyrimidin-4-yl)piperidin-4-yl)methylamine | | Descriptor: | 1-[4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-yl]methanamine, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
1Y0R
 
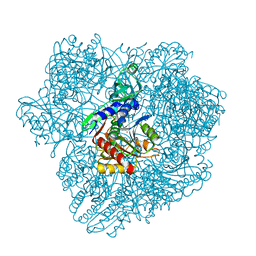 | |
8BNU
 
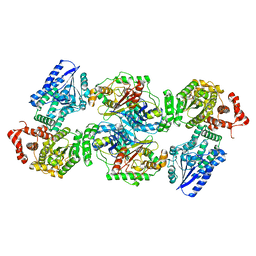 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) tetrameric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
4G73
 
 | | Crystal structure of NDH with NADH and Quinone | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, W, Feng, Y, Ge, J, Yang, M. | | Deposit date: | 2012-07-19 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
8BRJ
 
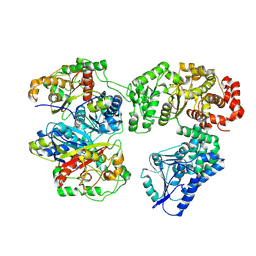 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) trimeric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
3M4J
 
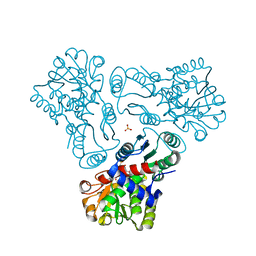 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible Post-Translational Carboxylation Modulates the Enzymatic Activity of N-Acetyl-l-ornithine Transcarbamylase.
Biochemistry, 49, 2010
|
|
2VNW
 
 | | Structure of PKA-PKB chimera complexed with (1-(9H-Purin-6-yl) piperidin-4-yl)methanamine | | Descriptor: | 1-[1-(9H-purin-6-yl)piperidin-4-yl]methanamine, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
2WC9
 
 | | Crystal structure of the g2p (large terminase) nuclease domain from the bacteriophage SPP1 with bound Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, TERMINASE LARGE SUBUNIT | | Authors: | Smits, C, Chechik, M, Kovalevskiy, O.V, Shevtsov, M.B, Foster, A.W, Alonso, J.C, Antson, A.A. | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Nuclease Activity of a Bacteriophage Large Terminase.
Embo Rep., 10, 2009
|
|
3LZO
 
 | | Crystal Structure Analysis of the copper-reconstituted P19 protein from Campylobacter jejuni at 1.65 A at pH 10.0 | | Descriptor: | COPPER (II) ION, P19 protein, SULFATE ION | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
2W1X
 
 | | The interdependence of wavelength, redundancy and dose in sulfur SAD experiments: 1.284 A wavelength 360 images data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
