5KR3
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5HLZ
 
 | |
1IEW
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 2-deoxy-2-fluoro-alpha-D-glucoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-fluoro-alpha-D-glucopyranose, ... | | Authors: | Hrmova, M, DeGori, R, Fincher, G.B, Smith, B.J, Varghese, J.N. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
1CI5
 
 | | GLYCAN-FREE MUTANT ADHESION DOMAIN OF HUMAN CD58 (LFA-3) | | Descriptor: | PROTEIN (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3(CD58)) | | Authors: | Sun, Z.Y.J, Dotsch, V, Kim, M, Li, J, Reinherz, E.L, Wagner, G. | | Deposit date: | 1999-04-07 | | Release date: | 1999-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Functional glycan-free adhesion domain of human cell surface receptor CD58: design, production and NMR studies.
EMBO J., 18, 1999
|
|
5KU0
 
 | | expanded poliovirus in complex with VHH 17B | | Descriptor: | VHH 17B, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
2Z7H
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with inhibitor BPH-210 | | Descriptor: | Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION, {1-HYDROXY-3-[METHYL(4-PHENYLBUTYL)AMINO]PROPANE-1,1-DIYL}BIS(PHOSPHONIC ACID) | | Authors: | Cao, R, Chen, C.K.-M, Guo, R.-T, Hudock, M, Wang, A.H.-J, Oldfield, E. | | Deposit date: | 2007-08-23 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of a potent phenylalkyl bisphosphonate inhibitor bound to farnesyl and geranylgeranyl diphosphate synthases.
Proteins, 73, 2008
|
|
1IQT
 
 | | Solution structure of the C-terminal RNA-binding domain of heterogeneous nuclear ribonucleoprotein D0 (AUF1) | | Descriptor: | heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Katahira, M, Miyanoiri, Y, Enokizono, Y, Matsuda, G, Nagata, T, Ishikawa, F, Uesugi, S. | | Deposit date: | 2001-08-01 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal RNA-binding domain of hnRNP D0 (AUF1), its interactions with RNA and DNA, and change in backbone dynamics upon complex formation with DNA.
J.Mol.Biol., 311, 2001
|
|
5HUO
 
 | | Crystal Structure of NadC Deletion Mutant in C2221 Space Group | | Descriptor: | Nicotinate-nucleotide diphosphorylase (Carboxylating), SULFATE ION | | Authors: | Booth, W.T, Chruszcz, M. | | Deposit date: | 2016-01-27 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
5L1D
 
 | |
1CIB
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH GDP, IMP, HADACIDIN, AND NO3 | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Hou, Z, Cashel, M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-03-31 | | Release date: | 2000-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase.
J.Biol.Chem., 274, 1999
|
|
5L5H
 
 | |
5L1C
 
 | |
5ULU
 
 | |
5HY0
 
 | | orotic acid hydrolase | | Descriptor: | GLYCEROL, Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5HYI
 
 | | Glycosylated, disulfide-linked Hole-Hole Fc fragment | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural differences between glycosylated, disulfide-linked heterodimeric Knob-into-Hole Fc fragment and its homodimeric Knob-Knob and Hole-Hole side products.
Protein Eng. Des. Sel., 30, 2017
|
|
5L5V
 
 | |
5L6B
 
 | | Yeast 20S proteasome with mouse beta5i (1-138) and mouse beta6 (97-111; 118-133) in complex with ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
3BKJ
 
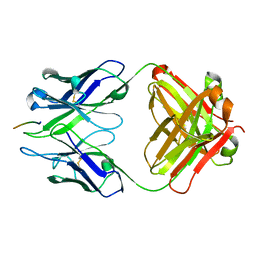 | | Crystal structure of Fab wo2 bound to the n terminal domain of amyloid beta peptide (1-16) | | Descriptor: | Amyloid Beta Peptide, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-06 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
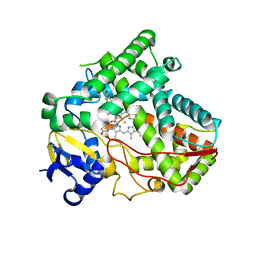
5VBU
 
 | |
1IIJ
 
 | |
1CPM
 
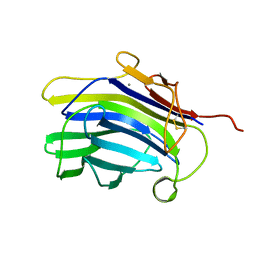 | |
5VPH
 
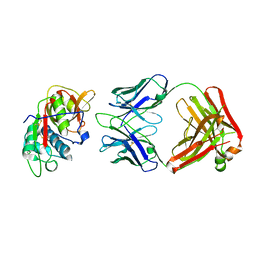 | | CRYSTAL STRUCTURE OF DER P 1 COMPLEXED WITH FAB 4C1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Der p 1 allergen, ... | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Determinants For Antibody Binding On Group 1 House Dust Mite Allergens.
J.Biol.Chem., 287, 2012
|
|
1CPY
 
 | | SITE-DIRECTED MUTAGENESIS ON (SERINE) CARBOXYPEPTIDASE Y FROM YEAST. THE SIGNIFICANCE OF THR 60 AND MET 398 IN HYDROLYSIS AND AMINOLYSIS REACTIONS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SERINE CARBOXYPEPTIDASE | | Authors: | Sorensen, S.B, Raaschou-Nielsen, M, Mortensen, U, Remington, S.J, Breddam, K. | | Deposit date: | 1995-03-24 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Site-Directed Mutagenesis on (Serine) Carboxypeptidase Y from Yeast. The Significance of Thr 60 and met 398 in Hydrolysis and Aminolysis Reactions
J.Am.Chem.Soc., 117, 1995
|
|
2PFS
 
 | | Crystal structure of universal stress protein from Nitrosomonas europaea | | Descriptor: | CHLORIDE ION, Universal stress protein | | Authors: | Chruszcz, M, Evdokimova, E, Cymborowski, M, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
1CSO
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-ILE18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
