6XUA
 
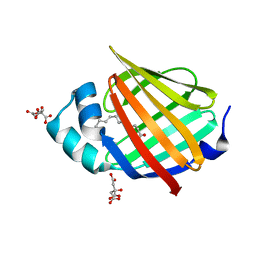 | | Human myelin protein P2 mutant K21Q | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
5IB3
 
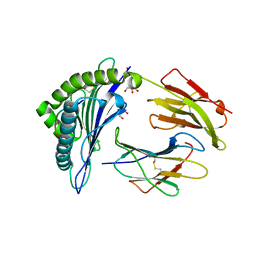 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR and Copper | | Descriptor: | Beta-2-microglobulin, COPPER (II) ION, GLYCEROL, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
7QEJ
 
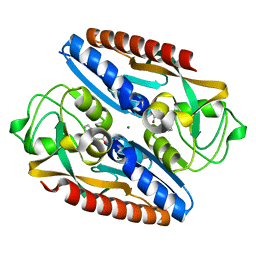 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-acetic acid (IAA). | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MAGNESIUM ION, TRANSCRIPTIONAL REGULATOR AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
6XVM
 
 | |
7QEK
 
 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-piruvic acid (IPA). | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, MAGNESIUM ION, regulator AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
3TJO
 
 | |
6XW9
 
 | | Human myelin protein P2 mutant K120S | | Descriptor: | CHLORIDE ION, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XWY
 
 | | Highly pH-resistant long stokes-shift, red fluorescent protein mCRISPRed | | Descriptor: | 1,2-ETHANEDIOL, MALONIC ACID, Red fluorescent protein eqFP611 | | Authors: | Erdogan, M, Fabritius, A, Basquin, J, Griesbeck, O. | | Deposit date: | 2020-01-24 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Targeted In Situ Protein Diversification and Intra-organelle Validation in Mammalian Cells.
Cell Chem Biol, 27, 2020
|
|
6XII
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-20 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
2YSV
 
 | | Solution structure of C2H2 type Zinc finger domain 17 in Zinc finger protein 473 | | Descriptor: | ZINC ION, Zinc finger protein 473 | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-04 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of C2H2 type Zinc finger domain 17 in Zinc finger protein 473
To be Published
|
|
4M3P
 
 | | Betaine-Homocysteine S-Methyltransferase from Homo sapiens complexed with Homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Betaine--homocysteine S-methyltransferase 1, POTASSIUM ION, ... | | Authors: | Koutmos, M, Yamada, K, Mladkova, J, Paterova, J, Diamond, C.E, Tryon, K, Jungwirth, P, Garrow, T.A, Jiracek, J. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Specific potassium ion interactions facilitate homocysteine binding to betaine-homocysteine S-methyltransferase.
Proteins, 82, 2014
|
|
3TLU
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-24' oxidized mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7QTR
 
 | | GB1 in mammalian cells, 50 uM | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Gerez, J.A, Prymaczok, N.C, Kadavath, H, Gosh, D, Butikofer, M, Guntert, P, Riek, R. | | Deposit date: | 2022-01-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in human cells by in-cell NMR and a reporter system to optimize protein delivery or transexpression.
Commun Biol, 5, 2022
|
|
7QTS
 
 | | GB1 in mammalian cells, 10 uM | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Gerez, J.A, Prymaczok, N.C, Kadavath, H, Gosh, D, Butikofer, M, Guntert, P, Riek, R. | | Deposit date: | 2022-01-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in human cells by in-cell NMR and a reporter system to optimize protein delivery or transexpression.
Commun Biol, 5, 2022
|
|
2YME
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht3 Receptors.
Embo Rep., 14, 2013
|
|
1HI2
 
 | | Eosinophil-derived Neurotoxin (EDN) - Sulphate Complex | | Descriptor: | EOSINOPHIL-DERIVED NEUROTOXIN, SULFATE ION | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI5
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
5OVK
 
 | | Crystal structure MabA bound to NADPH from M. smegmatis | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D8Q
 
 | |
2YS2
 
 | | Solution structure of the BTK motif of human Cytoplasmic tyrosine-protein kinase BMX | | Descriptor: | Cytoplasmic tyrosine-protein kinase BMX, ZINC ION | | Authors: | Abe, H, Tochio, N, Tomizawa, T, Koshiba, S, Yoneyama, M, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the BTK motif of human Cytoplasmic tyrosine-protein kinase BMX
To be Published
|
|
6XVN
 
 | |
6D8Y
 
 | |
5EX3
 
 | | Crystal structure of human SMYD3 in complex with a VEGFR1 peptide | | Descriptor: | ACETIC ACID, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Qiao, Q, Fu, W, Liu, N, Wang, M, Min, J, Zhu, B, Xu, R.M, Yang, N. | | Deposit date: | 2015-11-23 | | Release date: | 2016-03-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Structural Basis for Substrate Preference of SMYD3, a SET Domain-containing Protein Lysine Methyltransferase
J.Biol.Chem., 291, 2016
|
|
6D8R
 
 | |
7QNM
 
 | | Crystallization and structural analyses of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans | | Descriptor: | (S)-2-haloacid dehalogenase, PHOSPHATE ION | | Authors: | Grigorian, E, Roret, T, Leblanc, C, Delage, L, Czjzek, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | X-ray structure and mechanism of ZgHAD, a l-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 32, 2023
|
|
