7N97
 
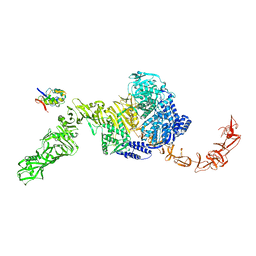 | | State 2 of TcdB and FZD2 at pH5 | | Descriptor: | Frizzled-2, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis for Receptor Recognition of the Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7N8X
 
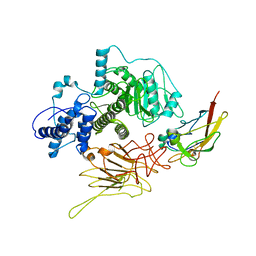 | | Partial C. difficile TcdB and CSPG4 fragment | | Descriptor: | Chondroitin sulfate proteoglycan 4, Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
7V9H
 
 | | The BEN3 domain of protein Bend3 | | Descriptor: | BEN domain-containing protein 3 | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
1ZZP
 
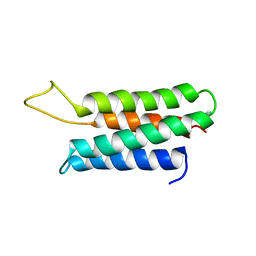 | | Solution structure of the F-actin binding domain of Bcr-Abl/c-Abl | | Descriptor: | Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Hantschel, O, Wiesner, S, Guttler, T, Mackereth, C.D, Rix, L.L.R, Mikes, Z, Dehne, J, Gorlich, D, Sattler, M, Superti-Furga, G. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Cytoskeletal Association of Bcr-Abl/c-Abl.
Mol.Cell, 19, 2005
|
|
224L
 
 | |
5IAX
 
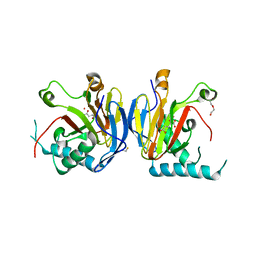 | |
5IBB
 
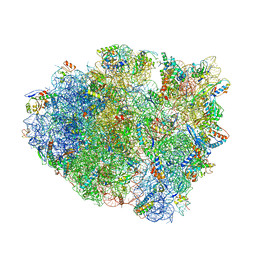 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and cognate tRNAVal in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The ribosome prohibits the GU wobble geometry at the first position of the codon-anticodon helix.
Nucleic Acids Res., 44, 2016
|
|
7V9F
 
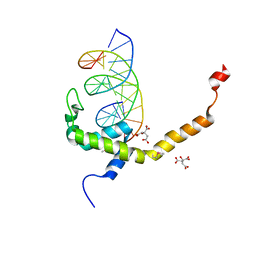 | | Selenomethionine mutant (L740Sem) of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, CITRIC ACID, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), ... | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7VGP
 
 | |
5IB8
 
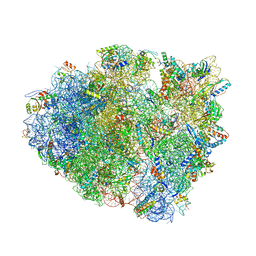 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and near-cognate tRNALys with U-G mismatch in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | The ribosome prohibits the GU wobble geometry at the first position of the codon-anticodon helix.
Nucleic Acids Res., 44, 2016
|
|
7VGO
 
 | | Hen egg lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Oda, M, Ikura, T, Ito, N. | | Deposit date: | 2021-09-17 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Hen Egg Lysozyme Refolded after Denaturation at Acidic pH.
Protein J., 41, 2022
|
|
5IMP
 
 | | Crystal structure of N299A Aspergillus terreus aristolochene synthase complexed with (1S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (1S,5S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Christianson, D.W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.038 Å) | | Cite: | Probing the Role of Active Site Water in the Sesquiterpene Cyclization Reaction Catalyzed by Aristolochene Synthase.
Biochemistry, 55, 2016
|
|
7N9Q
 
 | | State 3 of TcdB and FZD2 at pH5 | | Descriptor: | Toxin B | | Authors: | Jiang, M, Zhang, J. | | Deposit date: | 2021-06-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Receptor Recognition of Clostridium difficile Toxin B and its Dissociation upon Acidification
To Be Published
|
|
5IN2
 
 | |
264D
 
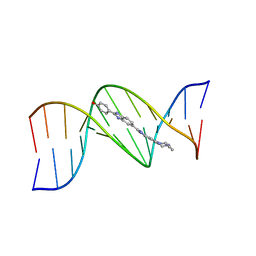 | | THREE-DIMENSIONAL CRYSTAL STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR-GROOVE-BINDING DRUG HOECHST 33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Vega, M.C, Garcia-Saez, I, Aymami, J, Eritja, R, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Coll, M. | | Deposit date: | 1994-09-22 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Three-dimensional crystal structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor-groove-binding drug Hoechst 33258.
Eur.J.Biochem., 222, 1994
|
|
7VFD
 
 | |
7VFG
 
 | |
5INT
 
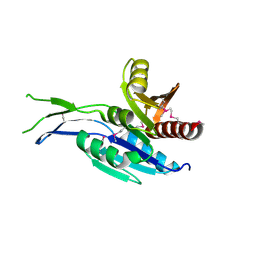 | | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC | | Descriptor: | Phosphopantothenate--cysteine ligase | | Authors: | Nocek, B, Zhou, M, Grimshaw, S, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-06 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC
To Be Published
|
|
2X99
 
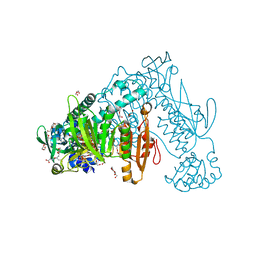 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with NADPH | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Angelucci, F, Dimastrogiovanni, D, Boumis, G, Brunori, M, Miele, A.E, Saccoccia, F, Bellelli, A. | | Deposit date: | 2010-03-15 | | Release date: | 2010-07-21 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping the Catalytic Cycle of Schistosoma Mansoni Thioredoxin Glutathione Reductase by X-Ray Crystallography
J.Biol.Chem., 285, 2010
|
|
1M6K
 
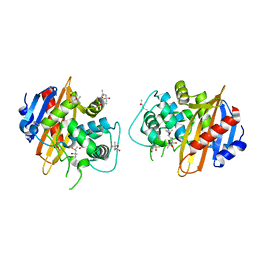 | | Structure of the OXA-1 class D beta-lactamase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, beta-lactamase OXA-1 | | Authors: | Sun, T, Nukaga, M, Mayama, K, Braswell, E.H, Knox, J.R. | | Deposit date: | 2002-07-16 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparison of beta-lactamases of classes A and D: 1.5A crystallographic structure of the class D OXA-1 oxacillinase
PROTEIN SCI., 12, 2003
|
|
7VFE
 
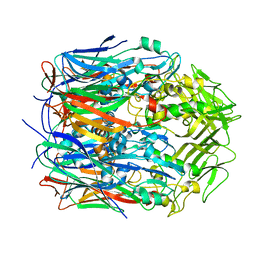 | |
7VFH
 
 | |
7VFF
 
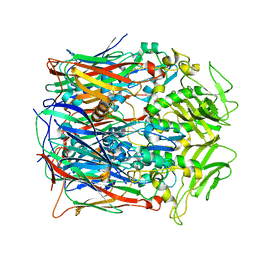 | |
7NFZ
 
 | |
2X72
 
 | | CRYSTAL STRUCTURE OF THE CONSTITUTIVELY ACTIVE E113Q,D2C,D282C RHODOPSIN MUTANT WITH BOUND GALPHACT PEPTIDE. | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ACETATE ION, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Standfuss, J, Edwards, P.C, Dantona, A, Fransen, M, Xie, G, Oprian, D.D, Schertler, G.F.X. | | Deposit date: | 2010-02-22 | | Release date: | 2011-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structural Basis of Agonist Induced Activation in Constitutively Active Rhodopsin
Nature, 471, 2011
|
|
