6Y1V
 
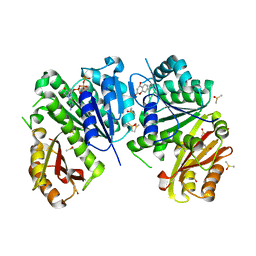 | | Mycobacterium tuberculosis FtsZ-GTP-gamma-S in complex with 4-hydroxycoumarin | | Descriptor: | 4-HYDROXY-2H-CHROMEN-2-ONE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein FtsZ, ... | | Authors: | Alnami, A.T, Norton, R.S, Pena, H.P, Haider, M, kozielski, F. | | Deposit date: | 2020-02-13 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Flexibility of A Highly Conserved Helix Controls Cryptic Pocket Formation in FtsZ.
J.Mol.Biol., 433, 2021
|
|
6Y1U
 
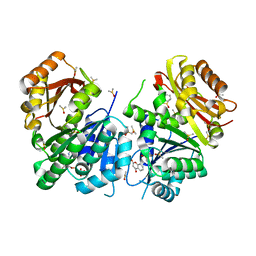 | | Mycobacterium tuberculosis FtsZ-GDP in complex with 4-hydroxycoumarin | | Descriptor: | 4-HYDROXY-2H-CHROMEN-2-ONE, Cell division protein FtsZ, DIMETHYL SULFOXIDE, ... | | Authors: | Alnami, A.T, Norton, R.S, Pena, H.P, Haider, M, kozielski, F. | | Deposit date: | 2020-02-13 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conformational Flexibility of A Highly Conserved Helix Controls Cryptic Pocket Formation in FtsZ.
J.Mol.Biol., 433, 2021
|
|
3PP6
 
 | | REP1-NXSQ fatty acid transporter Y128F mutant | | Descriptor: | PALMITOLEIC ACID, ReP1-NCXSQ | | Authors: | Berberian, G, Bollo, M, Howard, E, Cousido-Siah, A, Mitschler, A, Ayoub, D, Sanglier-Cianferani, S, Van Dorsselaer, A, DiPolo, R, Beauge, L, Petrova, T, Schulze-Briese, C, Wang, M, Podjarny, A. | | Deposit date: | 2010-11-24 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2MPB
 
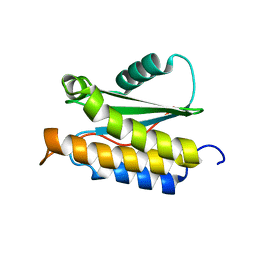 | | NMR structure of BA42 protein from the psychrophilic bacteria Bizionia argentinensis sp. nov | | Descriptor: | BA42 | | Authors: | Cicero, D, Aran, M, Smal, C, Pellizza, L, Gallo, M. | | Deposit date: | 2014-05-14 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution and crystal structure of BA42, a protein from the Antarctic bacterium Bizionia argentinensis comprised of a stand-alone TPM domain.
Proteins, 82, 2014
|
|
5TSF
 
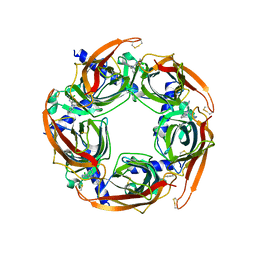 | | Crystal structure of AChBP from Aplysia californica complex with 2-aminopyrimidine at pH 7.0 spacegroup P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-N~4~,N~4~-bis[(pyridin-3-yl)methyl]pyrimidine-2,4-diamine, Soluble acetylcholine receptor | | Authors: | Camacho-Hernandez, G.A, Kaczanowska, K, Harel, M, Finn, M.G, Taylor, P.W. | | Deposit date: | 2016-10-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | Crystal structure of AChBP from Aplysia californica complex with 2-aminopyrimidine at pH 7.0 spacegroup P212121
To Be Published
|
|
6QD2
 
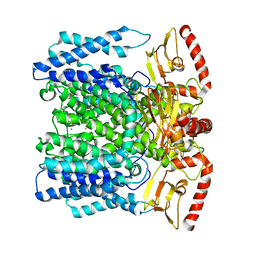 | | MloK1 model from single particle analysis of 2D crystals, class 6 (intermediate compact conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
2MVO
 
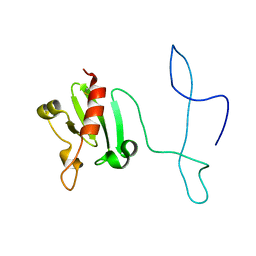 | | Solution structure of the lantibiotic self-resistance lipoprotein MlbQ from Microbispora ATCC PTA-5024 | | Descriptor: | Putative lipoprotein | | Authors: | Pozzi, R, Schwartz, P, Linke, D, Kulik, A, Nega, M, Wohlleben, W, Stegmann, E, Coles, M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Distinct mechanisms contribute to immunity in the lantibiotic NAI-107 producer strain Microbispora ATCC PTA-5024.
Environ Microbiol, 18, 2016
|
|
6BFV
 
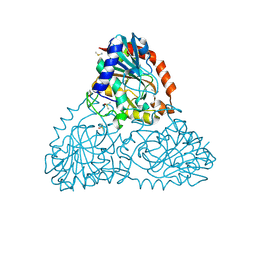 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 5-fluoro-1,2-dihydropyrimidin-2-one | | Descriptor: | 5-fluoropyrimidin-2-ol, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-10-27 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 5-fluoropyrimidin-1-ium-2-olate
To Be Published
|
|
7NMH
 
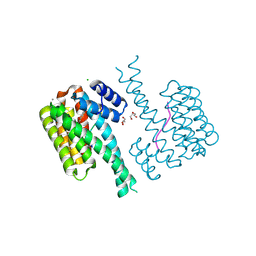 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-070 | | Descriptor: | (5-methanoyl-2-nitro-phenyl) methanesulfonate, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
3W1R
 
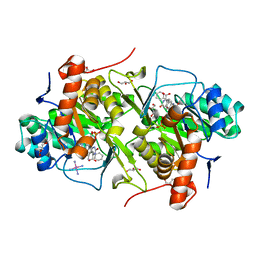 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-045a | | Descriptor: | 2,6-dioxo-5-(2-phenylethyl)-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-20 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-045a
To be Published
|
|
7KFZ
 
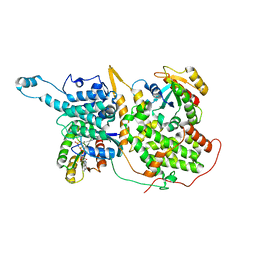 | | Structure of a ternary KRas(G13D)-SOS complex | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Liu, C, Moghadamchargari, Z, Laganowsky, A, Zhao, M. | | Deposit date: | 2020-10-15 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Molecular assemblies of the catalytic domain of SOS with KRas and oncogenic mutants.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1HJR
 
 | |
3W22
 
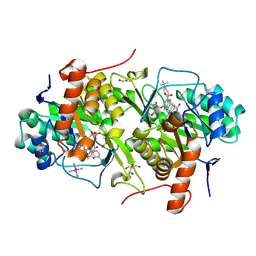 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-125 | | Descriptor: | 5-[2-(4-tert-butylphenyl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-125
To be Published
|
|
6PXM
 
 | | Horse spleen apoferritin light chain | | Descriptor: | Ferritin light chain | | Authors: | Kopylov, M, Kelley, K, Yen, L.Y, Rice, W.J, Eng, E.T, Carragher, B, Potter, C.S. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Horse spleen apoferritin light chain structure at 2.1 Angstrom resolution
To Be Published
|
|
3VOG
 
 | | Catalytic domain of the cellobiohydrolase, CcCel6A, from Coprinopsis cinerea | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cellobiohydrolase | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
6B63
 
 | | IMPase (AF2372) with 25 mM Asp | | Descriptor: | (2R)-2,3-dihydroxypropyl (2S)-2,3-dihydroxypropyl hydrogen phosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fructose-1,6-bisphosphatase/inositol-1-monophosphatase, ... | | Authors: | Goldstein, R.I, Roberts, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Osmolyte binding capacity of a dual action IMPase/FBPase (AF2372)
To Be Published
|
|
5NGS
 
 | | Crystal structure of human MTH1 in complex with inhibitor 6-[(2-phenylethyl)sulfanyl]-7H-purin-2-amine | | Descriptor: | 6-(2-phenylethylsulfanyl)-7~{H}-purin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
6B64
 
 | | IMPase (AF2372) with 25 mM Asp | | Descriptor: | ASPARTIC ACID, Fructose-1,6-bisphosphatase/inositol-1-monophosphatase, MAGNESIUM ION, ... | | Authors: | Goldstein, R.I, Roberts, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Osmolyte binding capacity of a dual action IMPase/FBPase (AF2372)
To Be Published
|
|
3VV0
 
 | | Crystal structure of histone methyltransferase SET7/9 in complex with DAAM-3 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl][2-(hexylamino)ethyl]amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase SETD7 | | Authors: | Niwa, H, Handa, N, Tomabechi, Y, Honda, K, Toyama, M, Ohsawa, N, Shirouzu, M, Kagechika, H, Hirano, T, Umehara, T, Yokoyama, S. | | Deposit date: | 2012-07-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structures of histone methyltransferase SET7/9 in complexes with adenosylmethionine derivatives
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6PXA
 
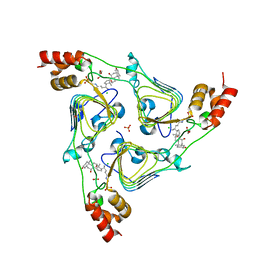 | | The crystal structure of chloramphenicol acetyltransferase-like protein from Vibrio fischeri ES114 in complex with taurocholic acid | | Descriptor: | ACETATE ION, CHLORIDE ION, Chloramphenicol acetyltransferase, ... | | Authors: | Tan, K, Maltseva, N, Jedrzejczak, R, Kuhn, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-25 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The crystal structure of chloramphenicol acetyltransferase-like protein from Vibrio fischeri ES114 in complex with taurocholic acid
To Be Published
|
|
5U42
 
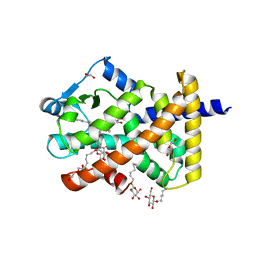 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 11 | | Descriptor: | 6-(2-{[cyclopropyl(3'-methoxy[1,1'-biphenyl]-4-carbonyl)amino]methyl}phenoxy)hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1REX
 
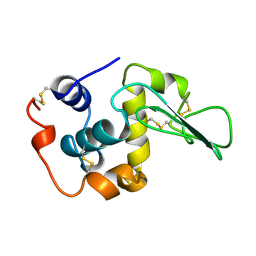 | | NATIVE HUMAN LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Muraki, M, Harata, K. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Origin of carbohydrate recognition specificity of human lysozyme revealed by affinity labeling.
Biochemistry, 35, 1996
|
|
6B82
 
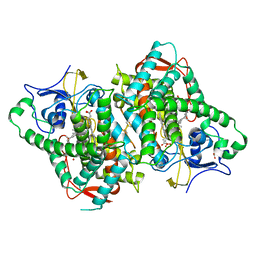 | | Zebra Fish CYP-450 17A1 Mutant Abiraterone Complex | | Descriptor: | ACETATE ION, Abiraterone, CHLORIDE ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-10-05 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Inherent steroid 17 alpha ,20-lyase activity in defunct cytochrome P450 17A enzymes.
J. Biol. Chem., 293, 2018
|
|
3VOS
 
 | | Crystal structure of Aspartate semialdehyde dehydrogenase Complexed With glycerol and sulfate From Mycobacterium tuberculosis H37Rv | | Descriptor: | Aspartate-semialdehyde dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Vyas, R, Tewari, R, Weiss, M.S, Karthikeyan, S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of ternary complexes of aspartate-semialdehyde dehydrogenase (Rv3708c) from Mycobacterium tuberculosis H37Rv
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5NGR
 
 | | Crystal structure of human MTH1 in complex with fragment inhibitor 8-(methylsulfanyl)-7H-purin-6-amine | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-methylsulfanyl-7~{H}-purin-6-amine, SULFATE ION | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
