7Y71
 
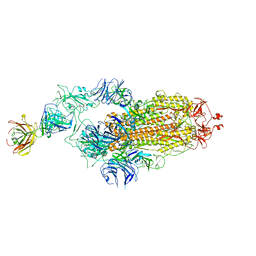 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab E7 heavy chain, ... | | Authors: | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | Deposit date: | 2022-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
6SR3
 
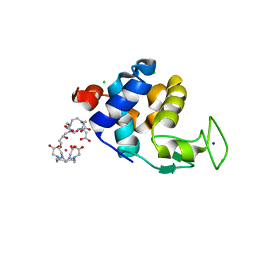 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 62 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
6SRA
 
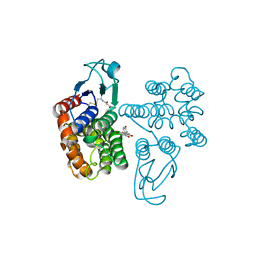 | | Crystal structure of glutathione transferase Omega 2C from Trametes versicolor in complex with naringenin | | Descriptor: | GLUTATHIONE, R-naringenin, glutathione transferase | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
1KN9
 
 | |
6IC5
 
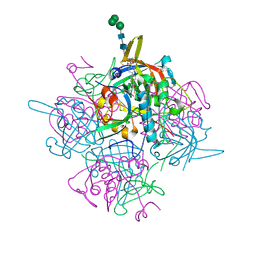 | | Human cathepsin-C in complex with dipeptidyl cyclopropyl nitrile inhibitor 2 | | Descriptor: | (2~{S})-2-azanyl-~{N}-[(1~{R},2~{R})-1-(iminomethyl)-2-[4-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]phenyl]cyclopropyl]-3-thiophen-2-yl-propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
6GHS
 
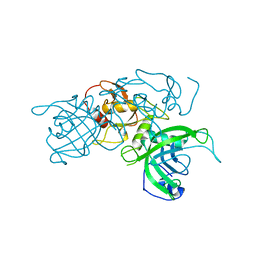 | | Modification dependent TagI restriction endonuclease | | Descriptor: | SODIUM ION, TagI restriction endonuclease, ZINC ION | | Authors: | Kisiala, M, Copelas, A, Czapinska, H, Xu, S, Bochtler, M. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal structure of the modification-dependent SRA-HNH endonuclease TagI.
Nucleic Acids Res., 46, 2018
|
|
1KJT
 
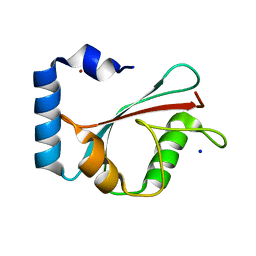 | | Crystal Structure of the GABA(A) Receptor Associated Protein, GABARAP | | Descriptor: | GABARAP, NICKEL (II) ION, SODIUM ION | | Authors: | Bavro, V.N, Sola, M, Bracher, A, Kneussel, M, Betz, H, Weissenhorn, W. | | Deposit date: | 2001-12-05 | | Release date: | 2002-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the GABA(A)-receptor-associated protein, GABARAP.
EMBO Rep., 3, 2002
|
|
6Y5V
 
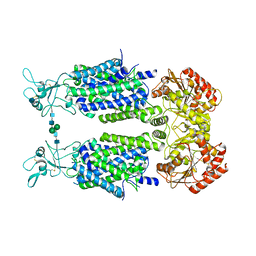 | | Structure of Human Potassium Chloride Transporter KCC3b (S45D/T940D/T997D) in KCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L. | | Deposit date: | 2020-02-25 | | Release date: | 2020-07-15 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
5GN6
 
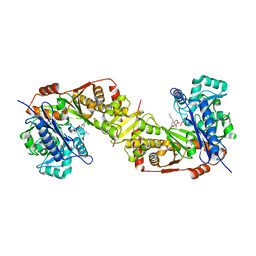 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17b | | Descriptor: | 4-butyl-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
20GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH CIBACRON BLUE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CIBACRON BLUE, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Nuccetelli, M, Mazzetti, A.P, Parker, M.W. | | Deposit date: | 1997-12-16 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
1CDN
 
 | |
6GKJ
 
 | | Translation initiation factor 4E in complex with trinucleotide mRNA 5' cap (m7GpppApG) | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2018-05-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Translation initiation factor 4E in complex with trinucleotide mRNA 5' cap (m7GpppApG)
To Be Published
|
|
5MCQ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE AND EXOSITE BINDING PEPTIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, BACE-1 ACTIVE AND EXOSITE BINDING INHIBITOR, Beta-secretase 1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
2ODT
 
 | | Structure of human Inositol 1,3,4-trisphosphate 5/6-kinase | | Descriptor: | Inositol-tetrakisphosphate 1-kinase | | Authors: | Busam, R.D, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Hammarstrom, M, Holmberg, S.L, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Sagemark, J, Sundstrom, M, Uppenberg, J, Van Den Berg, S, Weigelt, J, Persson, C, Thorsell, A.G, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-26 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of human Inositol 1,3,4-trisphosphate 5/6-kinase
To be Published
|
|
1KS0
 
 | | The First Fibronectin Type II Module from Human Matrix Metalloproteinase 2 | | Descriptor: | Matrix Metalloproteinase 2 | | Authors: | Gehrmann, M, Briknarova, K, Banyai, L, Patthy, L, Llinas, M. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The col-1 module of human matrix metalloproteinase-2 (MMP-2): structural/functional relatedness between gelatin-binding fibronectin type II modules and lysine-binding kringle domains.
Biol.Chem., 383, 2002
|
|
5B37
 
 | | Crystal structure of L-tryptophan dehydrogenase from Nostoc punctiforme | | Descriptor: | Tryptophan dehydrogenase | | Authors: | Wakamatsu, T, Sakuraba, H, Kitamura, M, Hakumai, Y, Ohnishi, K, Ashiuchi, M, Ohshima, T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Insights into l-Tryptophan Dehydrogenase from a Photoautotrophic Cyanobacterium, Nostoc punctiforme.
Appl. Environ. Microbiol., 83, 2017
|
|
6SZA
 
 | |
6GXP
 
 | | Cryo-EM structure of a rotated E. coli 70S ribosome in complex with RF3-GDPCP(RF3-only) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
5G1Y
 
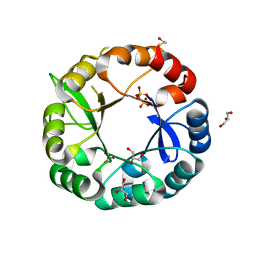 | | S. enterica HisA mutant D10G, dup13-15,V14:2M, Q24L, G102 | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-04-01 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ATH
 
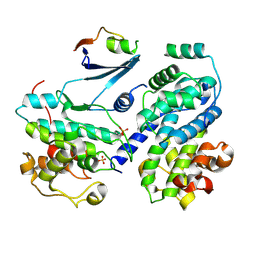 | | Cdk2/cyclin A/p27-KID-deltaC | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Dynamic anticipation by Cdk2/Cyclin A-bound p27 mediates signal integration in cell cycle regulation.
Nat Commun, 10, 2019
|
|
5G4E
 
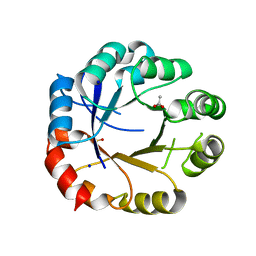 | | S. enterica HisA mutant D10G, Dup13-15, Q24L, G102A, V106L | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, ACETIC ACID, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M, Nasvall, J, Duarte, F, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-05-12 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5B4H
 
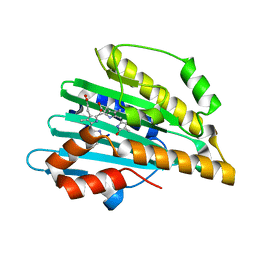 | | Crystal structure of I86D mutant of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin (data 1) | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Hagiwara, Y, Wada, K, Irikawa, T, Unno, M, Fukuyama, K, Sugishima, M. | | Deposit date: | 2016-04-04 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Atomic-resolution structure of the phycocyanobilin:ferredoxin oxidoreductase I86D mutant in complex with fully protonated biliverdin
FEBS Lett., 590, 2016
|
|
5ZHP
 
 | | M3 muscarinic acetylcholine receptor in complex with a selective antagonist | | Descriptor: | (1R,2R,4S,5S,7s)-7-({[4-fluoro-2-(thiophen-2-yl)phenyl]carbamoyl}oxy)-9,9-dimethyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-9-ium, CITRIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Liu, H, Hofmann, J, Fish, I, Schaake, B, Eitel, K, Bartuschat, A, Kaindl, J, Rampp, H, Banerjee, A, Hubner, H, Clark, M.J, Vincent, S.G, Fisher, J, Heinrich, M, Hirata, K, Liu, X, Sunahara, R.K, Shoichet, B.K, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6GGI
 
 | | Crystal structure of CotB2 in complex with 2-fluoro-3,7,18-dolabellatriene | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-fluoro-3,7,18-dolabellatriene, ACETONITRILE, ... | | Authors: | Driller, R, Janke, S, Fuchs, M, Warner, E, Mhashal, A.R, Major, D.T, Christmann, M, Brueck, T, Loll, B. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Towards a comprehensive understanding of the structural dynamics of a bacterial diterpene synthase during catalysis.
Nat Commun, 9, 2018
|
|
6IMU
 
 | | The apo-structure of endo-beta-1,2-glucanase from Talaromyces funiculosus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanaka, N, Nakajima, M, Narukawa-Nara, M, Matsunaga, H, Kamisuki, S, Aramasa, H, Takahashi, Y, Sugimoto, N, Abe, K, Miyanaga, A, Yamashita, T, Sugawara, F, Kamakura, T, Komba, S, Nakai, H, Taguchi, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and structural analyses of a fungal endo-beta-1,2-glucanase reveal a new glycoside hydrolase family.
J.Biol.Chem., 294, 2019
|
|
