1I0T
 
 | | 0.6 A STRUCTURE OF Z-DNA CGCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', SPERMINE | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.6 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
6VE7
 
 | | The inner junction complex of Chlamydomonas reinhardtii doublet microtubule | | Descriptor: | Cilia- and flagella-associated protein 20, FAP276, FAP52, ... | | Authors: | Khalifa, A.A.Z, Ichikawa, M, Bui, K.H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications.
Elife, 9, 2020
|
|
1V32
 
 | | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana | | Descriptor: | hypothetical protein RAFL09-47-K03 | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-24 | | Release date: | 2004-04-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana
To be Published
|
|
6UOQ
 
 | |
6M7M
 
 | | rac-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | L-GSTSTA from ice nucleation protein, inaZ, and its enantiomer, ... | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
2L7I
 
 | |
6M9T
 
 | | Crystal structure of EP3 receptor bound to misoprostol-FA | | Descriptor: | (11alpha,12alpha,13E,16S)-11,16-dihydroxy-16-methyl-9-oxoprost-13-en-1-oic acid, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, ... | | Authors: | Audet, M, White, K.L, Breton, B, Zarzycka, B, Han, G.W, Lu, Y, Gati, C, Batyuk, A, Popov, P, Velasquez, J, Manahan, D, Hu, H, Weierstall, U, Liu, W, Shui, W, Katrich, V, Cherezov, V, Hanson, M.A, Stevens, R.C. | | Deposit date: | 2018-08-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of misoprostol bound to the labor inducer prostaglandin E2receptor.
Nat. Chem. Biol., 15, 2019
|
|
1ULS
 
 | | Crystal structure of tt0140 from Thermus thermophilus HB8 | | Descriptor: | putative 3-oxoacyl-acyl carrier protein reductase | | Authors: | Hisanaga, Y, Ago, H, Hamada, K, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tt0140 from Thermus thermophilus HB8
To be Published
|
|
6V37
 
 | |
6V3I
 
 | |
6MAV
 
 | |
6V3Q
 
 | | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form | | Descriptor: | ISOPROPYL ALCOHOL, Metallo-beta-lactamase FIM-1, ZINC ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-26 | | Release date: | 2020-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form
To Be Published
|
|
1ZVJ
 
 | | Structure of Kumamolisin-AS mutant, D164N | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Nakayama, T. | | Deposit date: | 2005-06-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Processing, catalytic activity and crystal structures of kumamolisin-As with an engineered active site.
Febs J., 273, 2006
|
|
6UT1
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 LM/HS clade A/E CRF01 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
6V4P
 
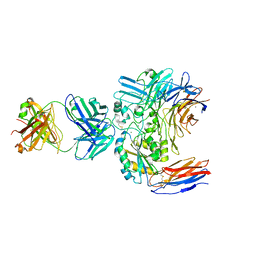 | | Structure of the integrin AlphaIIbBeta3-Abciximab complex | | Descriptor: | Abciximab, heavy chain, light chain, ... | | Authors: | Nesic, D, Zhang, Y, Spasic, A, Li, J, Provasi, D, Filizola, M, Walz, T, Coller, B.S. | | Deposit date: | 2019-11-28 | | Release date: | 2020-02-05 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-Electron Microscopy Structure of the alpha IIb beta 3-Abciximab Complex.
Arterioscler Thromb Vasc Biol., 40, 2020
|
|
1UUO
 
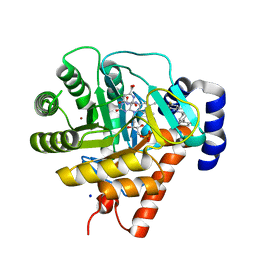 | | Rat dihydroorotate dehydrogenase (DHOD)in complex with brequinar | | Descriptor: | 6-FLUORO-2-(2'-FLUORO-1,1'-BIPHENYL-4-YL)-3-METHYLQUINOLINE-4-CARBOXYLIC ACID, DIHYDROOROTATE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hansen, M, Le Nours, J, Johansson, E, Antal, T, Ullrich, A, Loffler, M, Larsen, S. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Inhibitor Binding in a Class 2 Dihydroorotate Dehydrogenase Causes Variations in the Membrane-Associated N-Terminal Domain
Protein Sci., 13, 2004
|
|
6V76
 
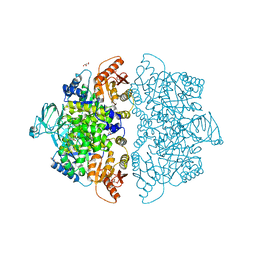 | | Crystal Structure of Human PKM2 in Complex with L-valine | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nandi, S, Dey, M. | | Deposit date: | 2019-12-07 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Biochemical and structural insights into how amino acids regulate pyruvate kinase muscle isoform 2.
J.Biol.Chem., 295, 2020
|
|
7AA4
 
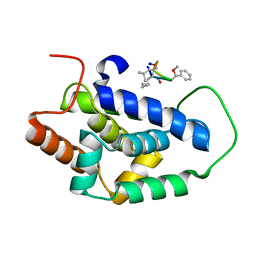 | | Structure of ClpC1-NTD bound to a CymA analogue | | Descriptor: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | Authors: | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
6V8H
 
 | | Crystal structure of Ara h 8.0201 | | Descriptor: | Ara h 8 allergen isoform, SULFATE ION | | Authors: | Offermann, L.R, Pote, S, Hurlburt, B.K, McBride, J.K, Chruszcz, M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of Ara h 8.0201
To Be Published
|
|
6MF9
 
 | | Crystal structure of CGD4-650 with compound BI2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, ZnKn (C2HC)+Athook+bromo domain protein, Taf250, ... | | Authors: | Dong, A, Lin, Y.L, Hou, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-10 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | Crystal structure of CGD4-650 with compound BI2536
to be published
|
|
1UZ0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with Glc-4Glc-3Glc-4Glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
6V54
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-03 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica.
To Be Published
|
|
1V63
 
 | | Solution structure of the 6th HMG box of mouse UBF1 | | Descriptor: | Nucleolar transcription factor 1 | | Authors: | Sato, M, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 6th HMG box of mouse UBF1
To be Published
|
|
6V6H
 
 | | Crystal structure of histidine ammonia-lyase from Trypanosoma cruzi | | Descriptor: | Histidine ammonia-lyase | | Authors: | Miranda, R.R, Silva, M, Barison, M.J, Silber, A.M, Iulek, J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of histidine ammonia-lyase from Trypanosoma cruzi.
Biochimie, 175, 2020
|
|
1VBO
 
 | | Crystal structure of artocarpin-mannotriose complex | | Descriptor: | alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, artocarpin | | Authors: | Jeyaprakash, A.A, Srivastav, A, Surolia, A, Vijayan, M. | | Deposit date: | 2004-02-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the carbohydrate specificities of artocarpin: variation in the length of a loop as a strategy for generating ligand specificity
J.Mol.Biol., 338, 2004
|
|
