2CEM
 
 | | P1' Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold | | Descriptor: | POL PROTEIN, {(1S)-1-[N'-[(2S)-2-HYDROXY-2-((1S,2R)-2-HYDROXY-INDAN-1-YLCARBAMOYL)-3-PHENYL-PROPYL]-N'-[4-(PYRIDINE-2-YL)-BENZYL]-HYDRAZINOCARBONYL]-2,2-DIMETHYL-PROPYL}-CARBAMIC ACID METHYL ESTER | | Authors: | Ginman, N, Ekegren, J.K, Johansson, A, Wallberg, H, Larhed, M, Samuelsson, B, Hallberg, A, Unge, T. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Microwave-Accelerated Synthesis of P1'-Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 49, 2006
|
|
2C75
 
 | | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins | | Descriptor: | AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE, N-PROPARGYL-1(S)-AMINOINDAN | | Authors: | Li, M, Binda, C, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2005-11-17 | | Release date: | 2006-04-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins
Biochemistry, 45, 2006
|
|
2CDE
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR | | Descriptor: | INKT-TCR | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
6DSO
 
 | | Cryo-EM structure of murine AA amyloid fibril | | Descriptor: | Serum amyloid A-2 protein | | Authors: | Loerch, S, Liberta, F, Grigorieff, N, Fandrich, M, Schmidt, M. | | Deposit date: | 2018-06-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids.
Nat Commun, 10, 2019
|
|
2CM0
 
 | |
1ZM7
 
 | | Crystal structure of D. melanogaster deoxyribonucleoside kinase mutant N64D in complex with dTTP | | Descriptor: | Deoxynucleoside kinase, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Welin, M, Skovgaard, T, Knecht, W, Berenstein, D, Munch-Petersen, B, Piskur, J, Eklund, H. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the changed substrate specificity of Drosophila melanogaster deoxyribonucleoside kinase mutant N64D.
Febs J., 272, 2005
|
|
5GJD
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 2 | | Descriptor: | 1-(4-((1H-pyrrolo[2,3-b]pyridin-4-yl)oxy)phenyl)-3-(5-(4-methylpiperazin-1-yl)naphthalen-2-yl)urea, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Development of a Method for Converting a TAK1 Type I Inhibitor into a Type II or c-Helix-Out Inhibitor by Structure-Based Drug Design (SBDD)
Chem.Pharm.Bull., 64, 2016
|
|
1Z1B
 
 | | Crystal structure of a lambda integrase dimer bound to a COC' core site | | Descriptor: | 26-MER DNA, 29-MER DNA, 5'-D(*CP*T*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
1ZN9
 
 | | Human Adenine Phosphoribosyltransferase in Apo and AMP Complexed Forms | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase | | Authors: | Iulek, J, Silva, M, Tomich, C.H.T.P, Thiemann, O.H. | | Deposit date: | 2005-05-11 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Complexes of Human Adenine Phosphoribosyltransferase Reveal Novel Features of the APRT Catalytic Mechanism
J.Biomol.Struct.Dyn., 25, 2008
|
|
6QSM
 
 | |
1ZO1
 
 | | IF2, IF1, and tRNA fitted to cryo-EM data OF E. COLI 70S initiation complex | | Descriptor: | P/I-site tRNA, translation Initiation Factor 1, translation initiation factor 2 | | Authors: | Allen, G.S, Zavialov, A, Gursky, R, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (13.8 Å) | | Cite: | The Cryo-EM Structure of a Translation Initiation Complex from Escherichia coli.
Cell(Cambridge,Mass.), 121, 2005
|
|
1Z6P
 
 | | Glycogen phosphorylase AMP site inhibitor complex | | Descriptor: | 4-{2-[(3-NITROBENZOYL)AMINO]PHENOXY}PHTHALIC ACID, Glycogen phosphorylase, muscle form | | Authors: | Kristiansen, M, Andersen, B, Iversen, L.F, Westergaard, N. | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification, synthesis and chracterization of new glycogen phosphorylase inhibitors binding to the allosteric AMP site
J.Med.Chem., 47, 2004
|
|
1Z8A
 
 | | Human Aldose Reductase complexed with novel Sulfonyl-pyridazinone Inhibitor | | Descriptor: | 6-[(5-CHLORO-3-METHYL-1-BENZOFURAN-2-YL)SULFONYL]PYRIDAZIN-3(2H)-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Podjarny, A, Heine, A, Klebe, G. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution crystal structure of aldose reductase complexed with the novel sulfonyl-pyridazinone inhibitor exhibiting an alternative active site anchoring group.
J.Mol.Biol., 356, 2006
|
|
1Z93
 
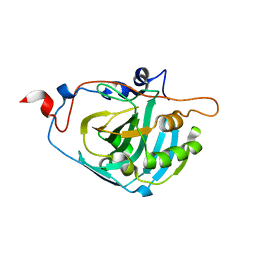 | | Human Carbonic Anhydrase III:Structural and Kinetic study of Catalysis and Proton Transfer. | | Descriptor: | Carbonic anhydrase III, ZINC ION | | Authors: | Duda, D.M, Tu, C, Fisher, S.Z, An, H, Yoshioka, C, Govindasamy, L, Laipis, P.J, Agbandje-McKenna, M, Silverman, D.N, McKenna, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer
Biochemistry, 44, 2005
|
|
1Z98
 
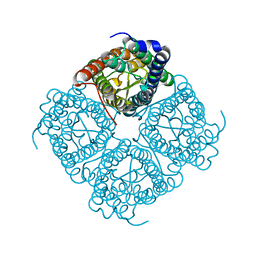 | | Crystal structure of the spinach aquaporin SoPIP2;1 in a closed conformation | | Descriptor: | CADMIUM ION, aquaporin | | Authors: | Tornroth-Horsefield, S, Hedfalk, K, Johanson, U, Karlsson, M, Neutze, R, Kjellbom, P. | | Deposit date: | 2005-04-01 | | Release date: | 2005-12-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural mechanism of plant aquaporin gating
Nature, 439, 2006
|
|
1Z9X
 
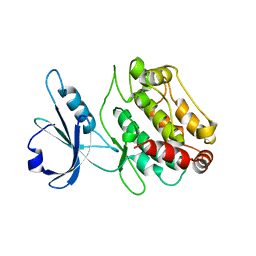 | | Human DRP-1 kinase, W305S S308A D40 mutant, crystal form with 3 monomers in the asymmetric unit | | Descriptor: | Death-associated protein kinase 2 | | Authors: | Kursula, P, Lehmann, F, Shani, G, Kimchi, A, Wilmanns, M. | | Deposit date: | 2005-04-05 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | A structural insight into the double-locking mechanism of the human death-associated DRP-1 kinase
To be Published
|
|
1ZAV
 
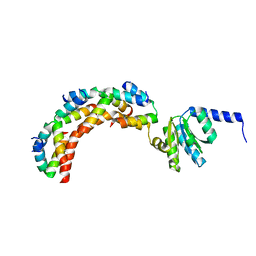 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P21 | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
1Z6Q
 
 | | Glycogen phosphorylase with inhibitor in the AMP site | | Descriptor: | 4-{2,4-BIS[(3-NITROBENZOYL)AMINO]PHENOXY}PHTHALIC ACID, Glycogen phosphorylase, muscle form | | Authors: | Kristiansen, M, Andersen, B, Iversen, L.F, Westergaard, N. | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification, synthesis and chracterization of new glycogen phosphorylase inhibitors binding to the allosteric AMP site
J.Med.Chem., 47, 2004
|
|
6HG8
 
 | | Crystal structure of the R460G disease-causing mutant of the human dihydrolipoamide dehydrogenase. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Ambrus, A, Szabo, E, Weichsel, A, Bui, D, Wilk, P, Torocsik, B, Weiss, M.S, Montfort, W.R, Jordan, F, Adam-Vizi, V. | | Deposit date: | 2018-08-22 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the R460G disease-causing mutant of the human dihydrolipoamide dehydrogenase.
To Be Published
|
|
1Z7M
 
 | | ATP Phosphoribosyl transferase (HisZG ATP-PRTase) from Lactococcus lactis | | Descriptor: | ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, PHOSPHATE ION, ... | | Authors: | Champagne, K.S, Sissler, M, Larrabee, Y, Doublie, S, Francklyn, C.S. | | Deposit date: | 2005-03-25 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation of the hetero-octameric ATP phosphoribosyl transferase through subunit interface rearrangement by a tRNA synthetase paralog.
J.Biol.Chem., 280, 2005
|
|
1Z89
 
 | | Human Aldose Reductase complexed with novel Sulfonyl-pyridazinone Inhibitor | | Descriptor: | 6-[(5-CHLORO-3-METHYL-1-BENZOFURAN-2-YL)SULFONYL]PYRIDAZIN-3(2H)-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Podjarny, A, Heine, A, Klebe, G. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | High-resolution crystal structure of aldose reductase complexed with the novel sulfonyl-pyridazinone inhibitor exhibiting an alternative active site anchoring group.
J.Mol.Biol., 356, 2006
|
|
5LO6
 
 | | HSP90 WITH indazole derivative | | Descriptor: | 3-(3,3-dimethylbutyl)-~{N}-methyl-~{N}-[4-(1-methylpiperidin-4-yl)phenyl]-6-oxidanyl-2~{H}-indazole-5-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Graedler, U, Amaral, M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
1ZF1
 
 | | CCC A-DNA | | Descriptor: | 5'-D(*CP*CP*GP*GP*GP*CP*CP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZFA
 
 | | GGA Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*TP*CP*CP*GP*GP*AP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZAL
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with partially disordered tagatose-1,6-bisphosphate, a weak competitive inhibitor | | Descriptor: | Fructose-bisphosphate aldolase A, PHOSPHATE ION | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
