3BJS
 
 | | Crystal structure of a member of enolase superfamily from Polaromonas sp. JS666 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Patskovsky, Y, Bonanno, J.B, Ozyurt, S, Dickey, M, Sauder, J.M, Reyes, C, Groshong, C, Gheyi, T, Smith, D, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Member of Enolase Superfamily from Polaromonas sp. JS666.
To be Published
|
|
7W4F
 
 | | Active state CI from Q1-NADH dataset, Subclass 4 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-27 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3B0Q
 
 | | Human PPAR gamma ligand binding domain in complex with MCC555 | | Descriptor: | (5S)-5-({6-[(2-fluorobenzyl)oxy]naphthalen-2-yl}methyl)-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-06-13 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human PPAR gamma in complex with MCC555
To be Published
|
|
7W62
 
 | | Mevo lectin complex with mannotetraose (Man4) | | Descriptor: | 1,2-ETHANEDIOL, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Sivaji, N, Vijayan, M. | | Deposit date: | 2021-12-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mevo lectin complex with mannotetraose (Man4)
To Be Published
|
|
3B0G
 
 | | Assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B1V
 
 | | Crystal structure of an S. thermophilus NFeoB E67A mutant bound to mGMPPNP | | Descriptor: | 3'-O-(N-methylanthraniloyl)-beta:gamma-imidoguanosine-5'-triphosphate, CHLORIDE ION, Ferrous iron uptake transporter protein B, ... | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-15 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A suite of Switch I and Switch II mutant structures from the G-protein domain of FeoB
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6I4T
 
 | | Crystal structure of the disease-causing I445M mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Wilk, P, Zambo, Z, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
7WNH
 
 | | Crystal structure of Nurr1 binding to NBRE | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*AP*AP*AP*GP*GP*TP*CP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*AP*TP*GP*AP*CP*CP*TP*TP*TP*TP*CP*GP*G)-3'), Nuclear receptor subfamily 4 group A member 2, ... | | Authors: | Zhao, M, Xu, T, Wang, N, Guo, Y, Liu, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Integrative analysis reveals structural basis for transcription activation of Nurr1 and Nurr1-RXR alpha heterodimer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7F3E
 
 | | Cryo-EM structure of the minimal protein-only RNase P from Aquifex aeolicus | | Descriptor: | RNA-free ribonuclease P | | Authors: | Teramoto, T, Koyasu, T, Adachi, N, Kawasaki, M, Moriya, T, Numata, T, Senda, T, Kakuta, Y. | | Deposit date: | 2021-06-16 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Minimal protein-only RNase P structure reveals insights into tRNA precursor recognition and catalysis.
J.Biol.Chem., 297, 2021
|
|
3B1Z
 
 | | Crystal structure of an S. thermophilus NFeoB T35S mutant without nucleotide | | Descriptor: | Ferrous iron uptake transporter protein B, SULFATE ION | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-15 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A suite of Switch I and Switch II mutant structures from the G-protein domain of FeoB
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7XZY
 
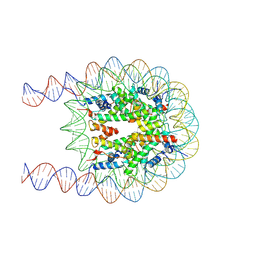 | | Cryo-EM structure of the nucleosome containing 193 base-pair DNA with a p53 target sequence | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
3B2L
 
 | | Iodide derivative of human LFABP | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
7Y12
 
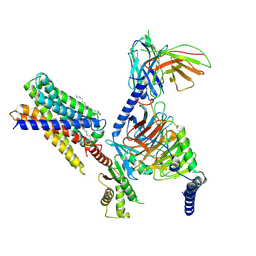 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine | | Descriptor: | BETA-ALANINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y14
 
 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine (local) | | Descriptor: | BETA-ALANINE, PALMITIC ACID, Soluble cytochrome b562,Mas-related G-protein coupled receptor member D | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7EQD
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOSPIRILLUM RUBRUM | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-azanyl-5-[(2~{E},6~{E},8~{E},10~{E},12~{E},14~{E},18~{E},22~{E},26~{E},30~{E},34~{E})-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,8,10,12,14,18,22,26,30,34,38-dodecaenyl]-3-methoxy-6-methyl-cyclohexa-2,5-diene-1,4-dione, CARDIOLIPIN, ... | | Authors: | Tani, K, Kanno, R, Ji, X.-C, Yu, L.-J, Hall, M, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-05-01 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM Structure of the Photosynthetic LH1-RC Complex from Rhodospirillum rubrum .
Biochemistry, 2021
|
|
7XTI
 
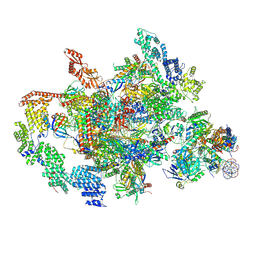 | | RNA polymerase II elongation complex transcribing a nucleosome (EC58hex) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7Y15
 
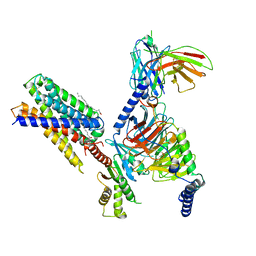 | | Cryo-EM structure of apo-state MrgD-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7XT7
 
 | | RNA polymerase II elongation complex transcribing a nucleosome (EC49B) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
3B6O
 
 | | Structure of TREX1 in complex with a nucleotide and an inhibitor ion (lithium) | | Descriptor: | LITHIUM ION, THYMIDINE-5'-PHOSPHATE, Three prime repair exonuclease 1 | | Authors: | Brucet, M, Querol-Audi, J, Fita, I, Celada, A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical studies of TREX1 inhibition by metals. Identification of a new active histidine conserved in DEDDh exonucleases.
Protein Sci., 17, 2008
|
|
7Y5Y
 
 | | X-ray Structure of Stay-Green (SGR) from Anaerolineae bacterium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Dey, D, Nishijima, M, Kurisu, G, Tanaka, H, Ito, H. | | Deposit date: | 2022-06-18 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and reaction mechanism of a bacterial Mg-dechelatase homolog from the Chloroflexi Anaerolineae.
Protein Sci., 31, 2022
|
|
3B46
 
 | |
7XZZ
 
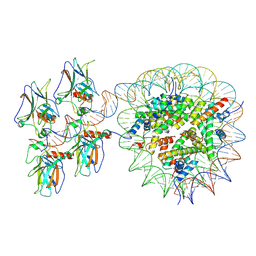 | | Cryo-EM structure of the nucleosome in complex with p53 | | Descriptor: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7F73
 
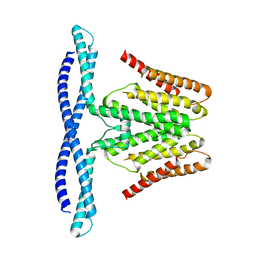 | | Cryo-EM structure of human TMEM120B | | Descriptor: | MCherry fluorescent protein,Transmembrane protein 120B | | Authors: | Ke, M, Wu, J, Yan, Z. | | Deposit date: | 2021-06-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of human TMEM120A and TMEM120B.
Cell Discov, 7, 2021
|
|
3B9U
 
 | |
3B73
 
 | |
