6YM5
 
 | | Crystal structure of BAY-091 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-fluoranyl-3-methyl-phenyl)phenyl]-1,7-naphthyridin-4-yl]amino]butanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
3CD5
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[3-(biphenyl-4-ylcarbamoyl)-2-ethyl-5,6,7,8-tetrahydrocyclohepta[b]pyrrol-1(4H)-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
6YI6
 
 | |
6YJL
 
 | |
6YJE
 
 | | Plasmoodium vivax phosphoglycerate kinase bound to nitrofuran inhibitor from PEG3350 and ammonium acetate at pH 5.5 | | Descriptor: | (2~{S})-2-(5-nitrofuran-2-yl)-2,3,5,6,7,8-hexahydro-1~{H}-[1]benzothiolo[2,3-d]pyrimidin-4-one, Phosphoglycerate kinase | | Authors: | Blaszczyk, B.K, Hyvonen, M. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Phosphoglycerate Kinase as a potential target for antimalarial therapy
to be published
|
|
6YMS
 
 | |
6YM3
 
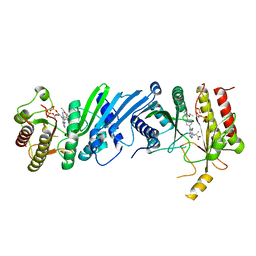 | | Crystal structure of Compound 1 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-ethoxyphenyl)phenyl]-5,8-dihydro-1,7-naphthyridin-4-yl]amino]propanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YM4
 
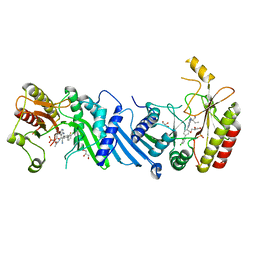 | | Crystal structure of BAY-297 with PIP4K2A | | Descriptor: | (2~{R})-2-[[2-[4-(3-chloranyl-2-fluoranyl-phenyl)phenyl]-3-cyano-1,7-naphthyridin-4-yl]amino]butanamide, GLYCEROL, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YMR
 
 | |
6YOU
 
 | |
6YPP
 
 | |
6YOT
 
 | |
6YPW
 
 | | Crystal structure for the complex of human carbonic anhydrase II and 4-((1-(2-(hydroxymethyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-1H-1,2,3-triazol-4-yl)methoxy)benzenesulfonamide | | Descriptor: | 4-[[1-[(2~{S},3~{S},5~{R})-2-(hydroxymethyl)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]oxolan-3-yl]-1,2,3-triazol-4-yl]methoxy]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2020-04-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mechanisms of the Antiproliferative and Antitumor Activity of Novel Telomerase-Carbonic Anhydrase Dual-Hybrid Inhibitors.
J.Med.Chem., 64, 2021
|
|
6YRT
 
 | |
6YS0
 
 | |
6YRM
 
 | |
6Y1T
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with a Trp51 to S-Trp51 modification | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2020-02-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
6Y2Y
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with Trp51 to S-Trp51 and Trp191Phe modifications | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2020-02-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
6YR6
 
 | | 14-3-3 sigma in complex with hDM2-186 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IODIDE ION, ... | | Authors: | Wolter, M, Srdanovic, S, Warriner, S, Wilson, A, Ottmann, C. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
6YR7
 
 | | 14-3-3 sigma in complex with hDMX-342+367 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein Mdm4 | | Authors: | Wolter, M, Srdanovic, S, Warriner, S, Wilson, A, Ottmann, C. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
6YR5
 
 | | 14-3-3 sigma in complex with hDMX-367 peptide | | Descriptor: | 14-3-3 protein sigma, Protein Mdm4, SULFATE ION | | Authors: | Wolter, M, Srdanovic, S, Ottman, C, Warriner, S, Wilson, A. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
6Y95
 
 | | Ca2+-free Calmodulin mutant N53I | | Descriptor: | Calmodulin | | Authors: | Holt, C, Hamborg, L.N, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
4OBQ
 
 | | MAP4K4 in complex with inhibitor (compound 31), N-[3-(4-AMINOQUINAZOLIN-6-YL)-5-FLUOROPHENYL]-2-(PYRROLIDIN-1-YL)ACETAMIDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Mitogen-activated protein kinase kinase kinase kinase 4, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-01-07 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of Selective 4-Amino-pyridopyrimidine Inhibitors of MAP4K4 Using Fragment-Based Lead Identification and Optimization.
J.Med.Chem., 57, 2014
|
|
6Y3K
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0403 | | Descriptor: | Major allergen variant Cor a 1.0403 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6YE5
 
 | | Structure of ribosomal binding factor A RbfA of Staphylococcus aureus bacterium by NMR | | Descriptor: | Ribosome-binding factor A | | Authors: | Blokhin, D.S, Usachev, K.S, Bikmullin, A.G, Nurullina, L, Garaeva, N, Validov, S, Klochkov, V, Aganov, A, Khusainov, I, Yusupov, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of ribosomal binding factor A RbfA of Staphylococcus aureus bacterium by NMR
To Be Published
|
|
