7Z4G
 
 | | SpCas9 bound to 12-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 12-nucleotide complementary DNA substrate, Target strand of 12-nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4K
 
 | | SpCas9 bound to 10-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 10-nucleotide complementary DNA substrate, Target strand of 10-nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4H
 
 | | SpCas9 bound to 14-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 14-nucleotide complementary DNA substrate, Target strand of 14-nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4J
 
 | |
7Z4L
 
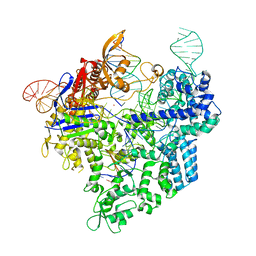 | |
3ELY
 
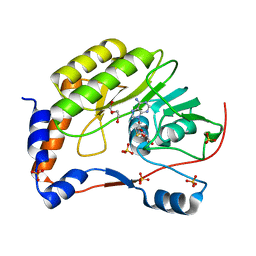 | | Wesselsbron virus Methyltransferase in complex with AdoHcy | | Descriptor: | DI(HYDROXYETHYL)ETHER, Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Bollati, M, Milani, M, Ricagno, S, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
3EMB
 
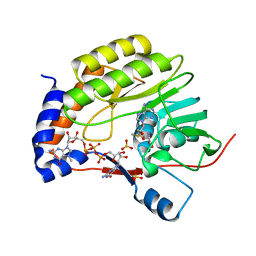 | | Wesselsbron virus Methyltransferase in complex with AdoMet and 7MeGpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Bollati, M, Ricagno, S, Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2008-09-24 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
3ETY
 
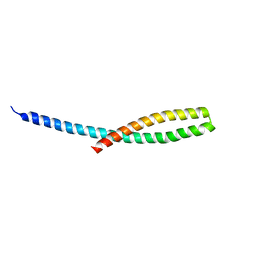 | | Crystal structure of bacterial adhesin FadA L14A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETW
 
 | | Crystal Structure of bacterial adhesin FadA | | Descriptor: | Adhesin A, THIOCYANATE ION | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3EBL
 
 | | Crystal Structure of Rice GID1 complexed with GA4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A4, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-08-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
3ELW
 
 | | Wesselsbron virus Methyltransferase in complex with AdoMet and GpppG | | Descriptor: | DIGUANOSINE-5'-TRIPHOSPHATE, Methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Bollati, M, Ricagno, S, Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
3ESX
 
 | | E16KE61KD126KD150K Flavodoxin from Anabaena | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Herguedas, B, Hermoso, J.A, Martinez-Julvez, M, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
3ESY
 
 | | E16KE61K Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
3ETX
 
 | | Crystal structure of bacterial adhesin FadA L14A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETZ
 
 | | Crystal structure of bacterial adhesin FadA L76A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
7QZ3
 
 | | The structure of T. forsythia NanH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
9C4I
 
 | | Centrolobium microchaete seed lectin (CML) complexed with Man1-3Man-OMe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Nascimento, K.S, Pinto-Junior, V.R, Lima, F.E.O, Osterne, V.J.S, Oliveira, M.V, Ferreira, V.M.S, Cavada, B.S. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Centrolobium microchaete seed lectin (CML) complexed with Man1-3Man-OMe
To Be Published
|
|
9AYC
 
 | |
2P6X
 
 | | Crystal structure of human tyrosine phosphatase PTPN22 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Ugochukwu, E, Salah, E, Barr, A, Shrestha, L, Alfano, I, Burgess-Brown, N, King, O, Umeano, C, Papagrigoriou, E, Pike, A.C.W, Bunkoczi, G, Turnbull, A, Uppenberg, J, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
6XYG
 
 | | Femtosecond structure of bovine trypsin at room temperature | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Jensen, M. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.50000238 Å) | | Cite: | High-resolution macromolecular crystallography at the FemtoMAX beamline with time-over-threshold photon detection
J.Synchrotron Radiat., 2021
|
|
6XZ2
 
 | |
2OOQ
 
 | | Crystal Structure of the Human Receptor Phosphatase PTPRT | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Receptor-type tyrosine-protein phosphatase T, ... | | Authors: | Ugochukwu, E, Alfano, I, Barr, A, Keates, T, Eswaran, J, Salah, E, Savitsky, P, Bunkoczi, G, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
6E8W
 
 | | MPER-TM Domain of HIV-1 envelope glycoprotein (Env) | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Fu, Q, Shaik, M.M, Cai, Y, Ghantous, F, Piai, A, Peng, H, Rits-Volloch, S, Liu, Z, Harrison, S.C, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane proximal external region of HIV-1 envelope glycoprotein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6XQW
 
 | | Crystal Structure of MaliM03 Fab in complex with Pfmsp1-19 | | Descriptor: | MaliM03 Fab Heavy Chain, MaliM03 Fab Light Chain, Pfmsp1-19 | | Authors: | Singh, S, Pancera, M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Multimeric antibodies from antigen-specific human IgM+ memory B cells restrict Plasmodium parasites.
J.Exp.Med., 218, 2021
|
|
6XUJ
 
 | | HumRadA1 in complex with 5-Ethyl-N-(1H-indol-5-ylmethyl)-1,3,4-thiadiazol-2-amine in P21212 | | Descriptor: | 5-Ethyl-N-(1H-indol-5-ylmethyl)-1,3,4-thiadiazol-2-amine, DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M.E, Scott, D.E, Hyvonen, M. | | Deposit date: | 2020-01-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To be published
|
|
