8RGG
 
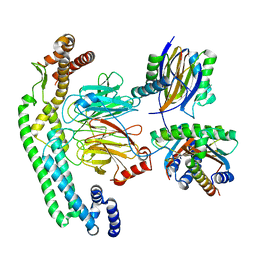 | | Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Dynein light chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Mladenov, M, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
5ZRG
 
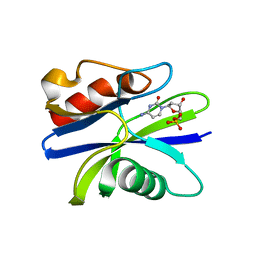 | | M. smegmatis antimutator protein MutT2 in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
8R0G
 
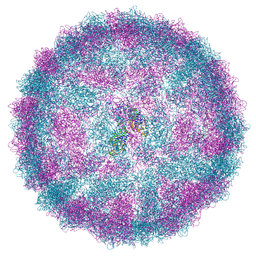 | | Capsid structure of Giardiavirus (GLV) CAT strain | | Descriptor: | Capsid protein | | Authors: | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Capsid structure of Giardiavirus (GLV) CAT strain
To Be Published
|
|
8R0F
 
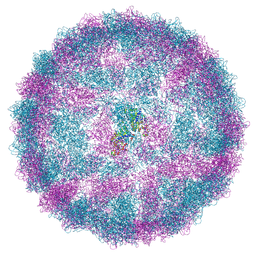 | | Capsid structure of Giardiavirus (GLV) HP strain | | Descriptor: | Capsid protein | | Authors: | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Capsid structure of Giardiavirus (GLV) HP strain
To Be Published
|
|
6A17
 
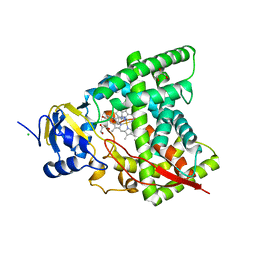 | | Crystal structure of CYP90B1 in complex with brassinazole | | Descriptor: | (2R,3S)-4-(4-chlorophenyl)-2-phenyl-3-(1H-1,2,4-triazol-1-yl)butan-2-ol, CHLORIDE ION, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
6P2S
 
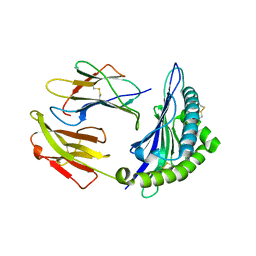 | |
5ZRK
 
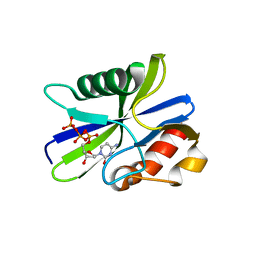 | | M. smegmatis antimutator protein MutT2 in complex with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZTE
 
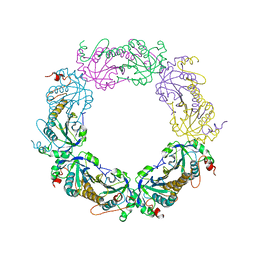 | | Crystal structure of PrxA C119S mutant from Arabidopsis thaliana | | Descriptor: | 2-Cys peroxiredoxin BAS1, chloroplastic | | Authors: | Yang, Y, Cai, W, Wang, J, Pan, W, Liu, L, Wang, M, Zhang, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Arabidopsis thaliana peroxiredoxin A C119S mutant.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6QBQ
 
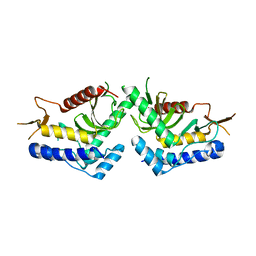 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S200A S203A | | Descriptor: | Cell wall assembly regulator SMI1 | | Authors: | Guillien, M, Batista, M, Francois, J.M, Mourey, L, Maveyraud, L, Zerbib, D. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S200A S203A
To Be Published
|
|
5ZYI
 
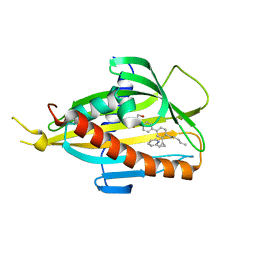 | | Crystal structure of CERT START domain in complex with compound E16 | | Descriptor: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
6QJ3
 
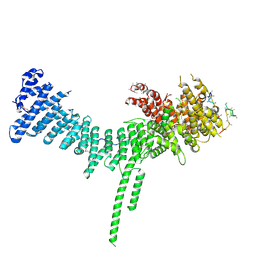 | | Crystal structure of the C. thermophilum condensin Ycs4-Brn1 subcomplex | | Descriptor: | Brn1, Condensin complex subunit 1,Condensin complex subunit 1,Ycs4, Condensin complex subunit 2 | | Authors: | Hassler, M, Haering, C.H, Kschonsak, M. | | Deposit date: | 2019-01-22 | | Release date: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of an Asymmetric Condensin ATPase Cycle.
Mol.Cell, 74, 2019
|
|
3BHD
 
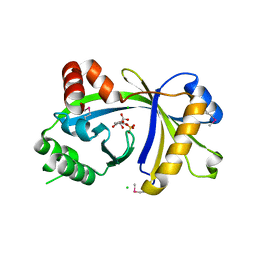 | | Crystal structure of human thiamine triphosphatase (THTPA) | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Busam, R.D, Lehtio, L, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Weigelt, J, Welin, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Thiamine Triphosphatase.
To be Published
|
|
7COJ
 
 | | Crystal structure of the b-carbonic anhydrase CafA of the fungal pathogen Aspergillus fumigatus | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Yeon, J, Sung, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of beta-Carbonic Anhydrase CafA from the Fungal Pathogen Aspergillus fumigatus .
Mol.Cells, 43, 2020
|
|
6QJA
 
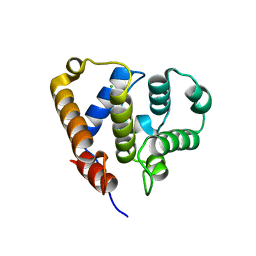 | | Organizational principles of the NuMA-Dynein interaction interface and implications for mitotic spindle functions | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nuclear mitotic apparatus protein 1 | | Authors: | Renna, C, Rizzelli, F, Carminati, M, Gaddoni, C, Pirovano, L, Cecatiello, V, Pasqualato, S, Mapelli, M. | | Deposit date: | 2019-01-23 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Organizational Principles of the NuMA-Dynein Interaction Interface and Implications for Mitotic Spindle Functions.
Structure, 28, 2020
|
|
6QGO
 
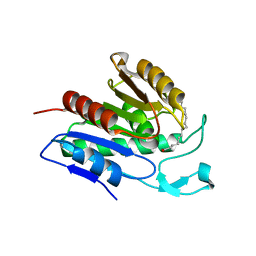 | | Crystal structure of APT1 S119A mutant bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
5ZXW
 
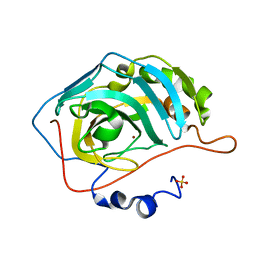 | | Crystal structure of human carbonic anhydrase II crystallized by ammonium sulfate | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION | | Authors: | Kitahara, M, Fudo, S, Yoneda, T, Nukaga, M, Hoshino, T. | | Deposit date: | 2018-05-22 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.316 Å) | | Cite: | Anisotropic Distribution of Ammonium Sulfate Ions in Protein Crystallization
Cryst.Growth Des., 19, 2019
|
|
6QF5
 
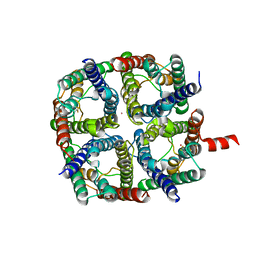 | | X-Ray structure of human Aquaporin 2 crystallized on a silicon chip | | Descriptor: | Aquaporin-2, CADMIUM ION | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
5ZYJ
 
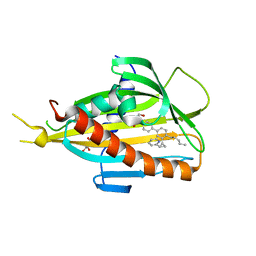 | | Crystal structure of CERT START domain in complex with compound E16A | | Descriptor: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZZE
 
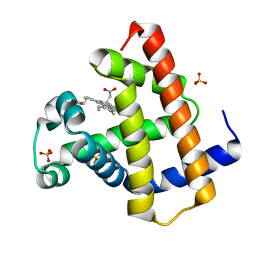 | | Crystal structure of horse myoglobin crystallized by ammonium sulfate | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kitahara, M, Fudo, S, Yoneda, T, Nukaga, M, Hoshino, T. | | Deposit date: | 2018-06-01 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Anisotropic Distribution of Ammonium Sulfate Ions in Protein Crystallization
Cryst.Growth Des., 19, 2019
|
|
6R73
 
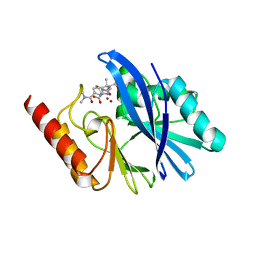 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, ZINC ION | | Authors: | Softley, C.A, Zak, K, Kolonko, M, Sattler, M, Popowicz, G. | | Deposit date: | 2019-03-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
7CW0
 
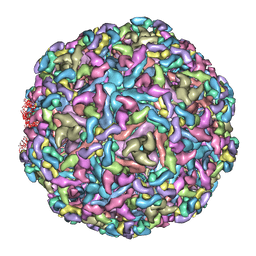 | | Cryo-EM structure of Chikungunya virus in complex with mAb CHK-263 IgG | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R78
 
 | | Structure of IMP-13 metallo-beta-lactamase in apo form (loop closed) | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
8TA5
 
 | | Title: Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with asymmetric C-terminal | | Descriptor: | Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
6A15
 
 | | Structure of CYP90B1 in complex with cholesterol | | Descriptor: | CHLORIDE ION, CHOLESTEROL, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
8TA4
 
 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with symmetric C-terminal | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
