6YQ5
 
 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - NMR Ensemble | | Descriptor: | Tail tube protein gp17.1* | | Authors: | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | Deposit date: | 2020-04-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
5N4W
 
 | | Crystal structure of the Cul2-Rbx1-EloBC-VHL ubiquitin ligase complex | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Cardote, T.A.F, Gadd, M.S, Ciulli, A. | | Deposit date: | 2017-02-11 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal Structure of the Cul2-Rbx1-EloBC-VHL Ubiquitin Ligase Complex.
Structure, 25, 2017
|
|
6YUC
 
 | | Crystal structure of Uba4-Urm1 from Chaetomium thermophilum | | Descriptor: | Adenylyltransferase and sulfurtransferase uba4, Ubiquitin-related modifier 1, ZINC ION | | Authors: | Grudnik, P, Pabis, M, Ethiraju Ravichandran, K, Glatt, S. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Molecular basis for the bifunctional Uba4-Urm1 sulfur-relay system in tRNA thiolation and ubiquitin-like conjugation.
Embo J., 39, 2020
|
|
6YXY
 
 | | State B of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
7OCJ
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS2(Nb14509) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, NbSOS2 (14509) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2021-04-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
8S03
 
 | | NMR solution structure of the CysD2 domain of MUC2 | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Recktenwald, C, Karlsson, B.G, Garcia-Bonnete, M.-J, Katona, G, Jensen, M, Lymer, R, Baeckstroem, M, Johansson, M.E.V, Hansson, G.C, Trillo-Muyo, S. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the second CysD domain of MUC2 and role in mucin organization by transglutaminase-based cross-linking.
Cell Rep, 43, 2024
|
|
4NAI
 
 | | Arabidopsis thaliana IspD apo | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, CADMIUM ION, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6YZN
 
 | | Closo-carborane butyl-sulfonamide in complex with CA IX mimic | | Descriptor: | Carbonic anhydrase 2, Carborane closo-butyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
6YZT
 
 | | Closo-carborane propyl-sulfonamide in complex with CA II | | Descriptor: | Carbonic anhydrase 2, Carborane propyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
6Z04
 
 | | Nido-carborane butyl-sulfonamide in complex with CA IX mimic | | Descriptor: | Carbonic anhydrase 2, Carborane nido-butyl-sulfonamide, ZINC ION | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
4NAK
 
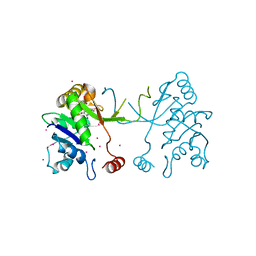 | | Arabidopsis thaliana IspD in complex with pentabromo-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5JD2
 
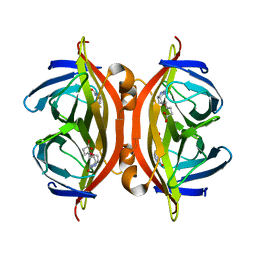 | | SFX structure of corestreptavidin-selenobiotin complex | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxohexahydro-1H-selenopheno[3,4-d]imidazol-4-yl]pentanoic acid, Streptavidin | | Authors: | DeMirci, H, Hunter, M.S, Boutet, S. | | Deposit date: | 2016-04-15 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selenium single-wavelength anomalous diffraction de novo phasing using an X-ray-free electron laser.
Nat Commun, 7, 2016
|
|
6Z6S
 
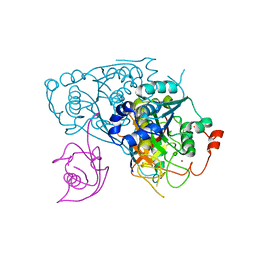 | | Crystal structure of Uba4-Urm1 from Chaetomium thermophilum | | Descriptor: | Adenylyltransferase and sulfurtransferase uba4, Ubiquitin-related modifier 1, ZINC ION | | Authors: | Grudnik, P, Pabis, M, Ethiraju Ravichandran, K, Glatt, S. | | Deposit date: | 2020-05-29 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Molecular basis for the bifunctional Uba4-Urm1 sulfur-relay system in tRNA thiolation and ubiquitin-like conjugation.
Embo J., 39, 2020
|
|
4NAN
 
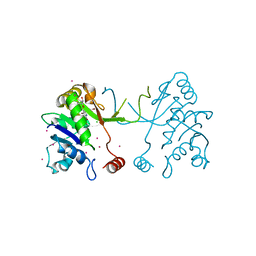 | | Arabidopsis thaliana IspD in complex with tetrabromo-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5J5Z
 
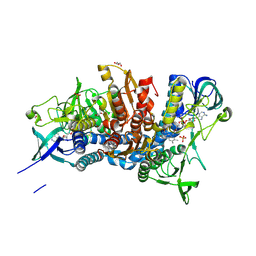 | | Crystal structure of the D444V disease-causing mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Mizsei, R, Zambo, Z, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2016-04-04 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of the disease-causing D444V mutant and the relevant wild type human dihydrolipoamide dehydrogenase.
Free Radic. Biol. Med., 124, 2018
|
|
1XD8
 
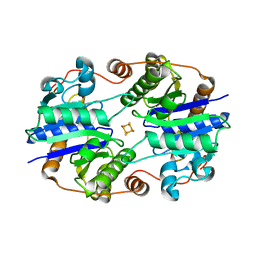 | | Crystal Structure of the Nitrogenase Fe protein Asp39Asn | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Jang, S.B, Jeong, M.S, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical implications of single amino acid substitutions in the nucleotide-dependent switch regions of the nitrogenase Fe protein from Azotobacter vinelandii
J.Biol.Inorg.Chem., 9, 2004
|
|
1XA1
 
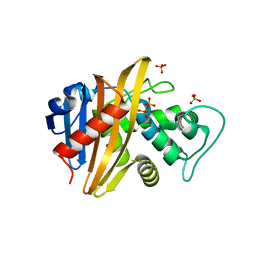 | | Crystal structure of the sensor domain of BlaR1 from Staphylococcus aureus in its apo form | | Descriptor: | PHOSPHATE ION, PYROPHOSPHATE 2-, Regulatory protein blaR1 | | Authors: | Wilke, M.S, Hills, T.L, Zhang, H.Z, Chambers, H.F, Strynadka, N.C. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
J.Biol.Chem., 279, 2004
|
|
6ZA4
 
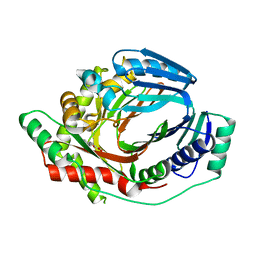 | | M. tuberculosis salicylate synthase MbtI in complex with 5-(3-cyanophenyl)furan-2-carboxylate | | Descriptor: | 5-(3-cyanophenyl)furan-2-carboxylic acid, CHLORIDE ION, Salicylate synthase | | Authors: | Mori, M, Villa, S, Meneghetti, F, Bellinzoni, M. | | Deposit date: | 2020-06-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Shedding X-ray Light on the Role of Magnesium in the Activity ofMycobacterium tuberculosisSalicylate Synthase (MbtI) for Drug Design.
J.Med.Chem., 63, 2020
|
|
1XCP
 
 | |
6YZS
 
 | | Carborane closo-pentyl-sulfonamide in complex with CA II | | Descriptor: | Carbonic anhydrase 2, Carborane closo-pentyl-sulfonamide, SODIUM ION, ... | | Authors: | Kugler, M, Brynda, J, Pospisilova, K, Rezacova, P. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The structural basis for the selectivity of sulfonamido dicarbaboranes toward cancer-associated carbonic anhydrase IX.
J Enzyme Inhib Med Chem, 35, 2020
|
|
1XE0
 
 | | The structure and function of Xenopus NO38-core, a histone binding chaperone in the nucleolus | | Descriptor: | Nucleophosmin | | Authors: | Namboodiri, V.M, Akey, I.V, Schmidt-Zachmann, M.S, Head, J.F, Akey, C.W. | | Deposit date: | 2004-09-08 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure and Function of Xenopus NO38-Core, a Histone Chaperone in the Nucleolus.
Structure, 12, 2004
|
|
1X78
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-244 | | Descriptor: | Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1, [5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-YL]ACETONITRILE | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
7OLZ
 
 | | Crystal structure of the SARS-CoV-2 RBD with neutralizing-VHHs Re5D06 and Re9F06 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Nanobody Re5D06, Nanobody Re9F06, ... | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
5M4G
 
 | | Crystal Structure of Wild-Type Human Prolidase with Mn ions | | Descriptor: | GLYCEROL, HYDROXIDE ION, MANGANESE (II) ION, ... | | Authors: | Wilk, P, Weiss, M.S, Mueller, U, Dobbek, H. | | Deposit date: | 2016-10-18 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Substrate specificity and reaction mechanism of human prolidase.
FEBS J., 284, 2017
|
|
8SVE
 
 | |
