7OOZ
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzyloxo-2-chloropurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzyloxo-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7OOY
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzylthio-2-chloropurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzylthio-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7OP9
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 2,6-dichloropurine | | Descriptor: | 2,6-bis(chloranyl)-7H-purine, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
4RBZ
 
 | |
7OPA
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzylthiopurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzylthio-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
1E3Q
 
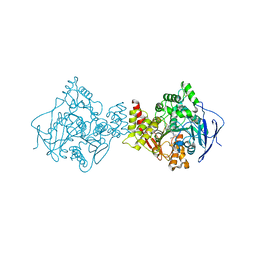 | | TORPEDO CALIFORNICA ACETYLCHOLINESTERASE COMPLEXED WITH BW284C51 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(5-{4-[DIMETHYL(PROP-2-ENYL)AMMONIO]PHENYL}-3-OXOPENTYL)-N,N-DIMETHYL-N-PROP-2-ENYLBENZENAMINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Felder, C.E, Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 2000-06-21 | | Release date: | 2000-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of a Complex of the Potent and Specific Inhibitor Bw284C51 with Torpedo Californica Acetylcholinesterase
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7OYJ
 
 | |
8HPT
 
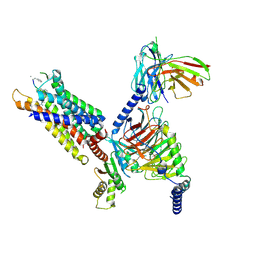 | | Structure of C5a-pep bound mouse C5aR1 in complex with Go | | Descriptor: | Antibody fragment ScFv16, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saha, S, Maharana, J, Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8KDU
 
 | | Crystal structure of trypsin with its substrate | | Descriptor: | (2~{R})-2-benzamido-5-carbamimidamido-pentanoic acid, ARGININE, CALCIUM ION, ... | | Authors: | Akbar, Z, Ahmad, M.S. | | Deposit date: | 2023-08-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Michaelis complex of trypsin with N-alpha-benzoyl-l-arginine ethyl ester
J Chin Chem Soc, n/a, 2024
|
|
4R6V
 
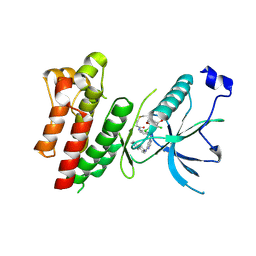 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Harboring the V550L Gate-Keeper Mutation in Complex with FIIN-3, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-[4-({[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino}methyl)phenyl]propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4R6Z
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-ETPP, Cs+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CESIUM ION, GLYCINE, ... | | Authors: | De March, M, Napolitano, L.M.R, Onesti, S. | | Deposit date: | 2014-08-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1PF7
 
 | | CRYSTAL STRUCTURE OF HUMAN PNP COMPLEXED WITH IMMUCILLIN H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | De Azevedo Jr, W.F, Canduri, F, Dos Santos, D.M, Pereira, J.H, Dias, M.V.B, Silva, R.G, Mendes, M.A, Palma, M.S, Basso, L.A, Santos, D.S. | | Deposit date: | 2003-05-24 | | Release date: | 2004-06-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for inhibition of human PNP by immucillin-H
Biochem.Biophys.Res.Commun., 309, 2003
|
|
8HFP
 
 | | Crystal structure of the methyl-CpG-binding domain of SETDB2 in complex with the cysteine-rich domain of C11orf46 protein | | Descriptor: | ARL14 effector protein, Histone-lysine N-methyltransferase SETDB2, SULFATE ION, ... | | Authors: | Mahana, Y, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural evidence for protein-protein interaction between the non-canonical methyl-CpG-binding domain of SETDB proteins and C11orf46.
Structure, 32, 2024
|
|
4R8C
 
 | | Crystal Structure of CNG mimicking NaK-ETPP mutant in complex with Rb+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, Potassium channel protein, ... | | Authors: | De March, M, Napolitano, L.M.R, Onesti, S. | | Deposit date: | 2014-09-01 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7OSO
 
 | | The crystal structure of Erwinia tasmaniensis levansucrase in complex with (S)-1,2,4-butanentriol | | Descriptor: | (2~{S})-butane-1,2,4-triol, Levansucrase (Beta-D-fructofuranosyl transferase), ZINC ION | | Authors: | Polsinelli, I, Salomone-Stagni, M, Benini, S. | | Deposit date: | 2021-06-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Erwinia tasmaniensis levansucrase shows enantiomer selection for (S)-1,2,4-butanetriol.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7P98
 
 | | Cyclohex-1-ene-1-carboxyl-CoA dehydrogenase in a substrate-free state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Short-chain acyl-CoA dehydrogenase | | Authors: | Ermler, U, Weidenweber, S, Boll, M. | | Deposit date: | 2021-07-26 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Cyclic 1,3-Diene Forming Acyl-Coenzyme A Dehydrogenases.
Chembiochem, 22, 2021
|
|
7P9X
 
 | | Structure of cyclohex-1-ene-1-carboxyl-CoA dehydrogenase complexed with cyclohex-1-ene-1-carboxyl-CoA | | Descriptor: | 1-monoenoyl-CoA, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ermler, U, Weidenweber, S, Boll, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Cyclic 1,3-Diene Forming Acyl-Coenzyme A Dehydrogenases.
Chembiochem, 22, 2021
|
|
1DME
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1DMC
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1DMD
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
4RAI
 
 | | Crystal Structure of CNG mimicking NaK-ETPP mutant in complex with Na+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, Potassium channel protein, ... | | Authors: | De March, M, Napolitano, L.M.R, Onesti, S. | | Deposit date: | 2014-09-10 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7P9A
 
 | | Structure of cyclohex-1-ene-1-carboxyl-CoA dehydrogenase complexed with cyclohex-1,5-diene-1-carboxyl-CoA | | Descriptor: | 1,5 Dienoyl-CoA, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ermler, U, Weidenweber, S, Boll, M. | | Deposit date: | 2021-07-26 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Cyclic 1,3-Diene Forming Acyl-Coenzyme A Dehydrogenases.
Chembiochem, 22, 2021
|
|
1DCL
 
 | |
7TN9
 
 | | Structure of the Inmazeb cocktail and resistance to escape against Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, GP2, ... | | Authors: | Rayaprolu, V, Fulton, B, Rafique, A, Arturo, E, Williams, D, Hariharan, C, Callaway, H, Parvate, A, Schendel, S.L, Parekh, D, Hui, S, Shaffer, K, Pascal, K.E, Wloga, E, Giordano, S, Copin, R, Franklin, M, Boytz, R.M, Donahue, C, Davey, R, Baum, A, Kyratsous, C.A, Saphire, E.O. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the Inmazeb cocktail and resistance to Ebola virus escape.
Cell Host Microbe, 31, 2023
|
|
8HW1
 
 | | Far-red light-harvesting complex of Antarctic alga Prasiola crispa | | Descriptor: | (1S)-4-[(1E,3Z,5E,7E,9E,11E,13E,15E,17E)-3-(hydroxymethyl)-7,12,16-trimethyl-18-[(1R,4S)-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]-3,5,5-trimethyl-cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, CHLOROPHYLL A, ... | | Authors: | Kosugi, M, Kawasaki, M, Shibata, Y, Moriya, T, Adachi, N, Senda, T. | | Deposit date: | 2022-12-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Uphill energy transfer mechanism for photosynthesis in an Antarctic alga.
Nat Commun, 14, 2023
|
|
