9AZI
 
 | |
9MHT
 
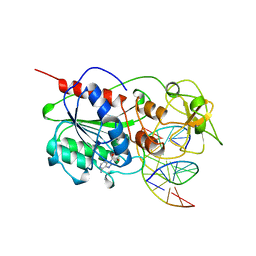 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*(3DR)P*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-07 | | Release date: | 1998-12-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
9FST
 
 | | Yeast 20S proteasome with human beta1i (1-51) in complex with epoxyketone inhibitor LU-001i | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
To Be Published
|
|
9FLV
 
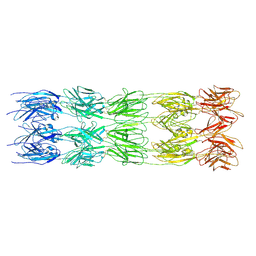 | | CryoEM structure of the fragment-5 (2393-2807) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9FLP
 
 | | CryoEM structure of the neck (1403-2314) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
4Q3O
 
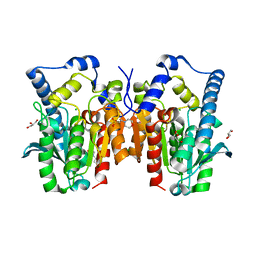 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
6QGB
 
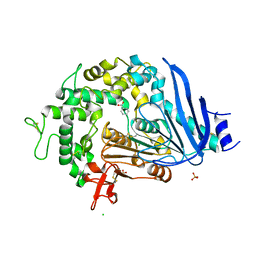 | | Crystal structure of Ideonella sakaiensis MHETase bound to benzoic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZOIC ACID, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
9FLX
 
 | | CryoEM structure of the fragment-4 (4074-4421) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9AXS
 
 | |
9FT1
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 13 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
1D8U
 
 | | CRYSTAL STRUCTURE OF NON-SYMBIOTIC PLANT HEMOGLOBIN FROM RICE | | Descriptor: | NON-SYMBIOTIC HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, M, Brucker, E.A, Stec, B, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1999-10-26 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a nonsymbiotic plant hemoglobin.
Structure Fold.Des., 8, 2000
|
|
9AXP
 
 | |
9G8B
 
 | | CryoEM structure of the fragment-2 (2892-3236) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9CSY
 
 | | SARS-CoV-2 papain-like protease (PLpro) bound to PF-07957472 | | Descriptor: | 2-methyl-5-(4-methylpiperazin-1-yl)-N-{1-[(2P)-2-(1-methyl-1H-pyrazol-4-yl)quinolin-4-yl]cyclopropyl}benzamide, Papain-like protease, ZINC ION, ... | | Authors: | Mashalidis, E.H, Chang, J.S, Wu, H, Garnsey, M. | | Deposit date: | 2024-07-24 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Discovery of SARS-CoV-2 papain-like protease (PL pro ) inhibitors with efficacy in a murine infection model.
Sci Adv, 10, 2024
|
|
9FLW
 
 | | CryoEM structure of the fragment-6 (3571-4078) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9FLS
 
 | | CryoEM structure of the fragment-3 (2061-2397) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9FLY
 
 | | CryoEM structure of the base (4151-5009) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
8G49
 
 | | FphE, Staphylococcus aureus fluorophosphonate-binding serine hydrolases E, Oxadiazolone compound 3 bound | | Descriptor: | Fluorophosphonate-binding serine hydrolase E, methyl 2-formyl-2-[3-methyl-4-(3-phenoxybenzamido)phenyl]hydrazine-1-carboxylate | | Authors: | Fellner, M, Bakker, A.T, Martin, N.I, Stelt, M. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | FphE, Staphylococcus aureus fluorophosphonate-binding serine hydrolases E, Oxadiazolone compound 3 bound
To be published
|
|
8QQJ
 
 | | CryoEM structure of the type IV pilin PilA5 from Thermus thermophilus | | Descriptor: | 7-Acetamido-5-acetimidoyl-3,5,7,9-tetradeoxy-L-glycero-L-manno-nonulosonic aci-(1-4)-alpha-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, MAGNESIUM ION, Type IV narrow pilus major component PilA5 | | Authors: | Gold, V.A.M, Neuhaus, A, Gaines, M, Isupov, M, McLaren, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | CryoEM structure of the type IV pilin PilA4 from Thermus thermophilus
To Be Published
|
|
9FT0
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 42 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
7SQO
 
 | | Structure of the orexin-2 receptor(OX2R) bound to TAK-925, Gi and scFv16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | McGrath, A.P, Kang, Y, Flinspach, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
8QT8
 
 | | Crystal structure of human Sirt2 in complex with a TNFa-Myr analogue TNFn-34 | | Descriptor: | 3-dodecylsulfanylpropanoic acid, NAD-dependent protein deacetylase sirtuin-2, Peptide-based TNFa-Myr analogue TNFn-34, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
7NWY
 
 | |
7O2S
 
 | | Crystal structure of a tetrameric form of Carbonic anhydrase from Schistosoma mansoni | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Carbonic anhydrase, ... | | Authors: | Ferraroni, M, Angeli, A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Insights into Schistosoma mansoni Carbonic Anhydrase (SmCA) Inhibition by Selenoureido-Substituted Benzenesulfonamides.
J.Med.Chem., 64, 2021
|
|
8JEE
 
 | | Crystal Structure of Human Carbonic Anhydrase II In-complex with Levosulpiride at 2.96 A Resolution | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, Levosulpiride, ... | | Authors: | Rasheed, S, Huda, N, Falke, S, Fisher, S.Z, Ahmad, M.S. | | Deposit date: | 2023-05-15 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal Structure of Human Carbonic Anhydrase II In-complex with Levosulpiride at 2.96 A Resolution
To Be Published
|
|
