3U6P
 
 | | MutM set 1 GpG | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*CP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*GP*GP*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
2Z0B
 
 | | Crystal structure of CBM20 domain of human putative glycerophosphodiester phosphodiesterase 5 (KIAA1434) | | Descriptor: | PHOSPHATE ION, Putative glycerophosphodiester phosphodiesterase 5 | | Authors: | Saijo, S, Nishino, A, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CBM20 domain of human putative glycerophosphodiester phosphodiesterase 5 (KIAA1434)
To be Published
|
|
3GOY
 
 | | Crystal structure of human poly(adp-ribose) polymerase 14, catalytic fragment in complex with an inhibitor 3-aminobenzamide | | Descriptor: | 3-aminobenzamide, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Moche, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
2HTE
 
 | | The crystal structure of spermidine synthase from p. falciparum in complex with 5'-methylthioadenosine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, SULFATE ION, ... | | Authors: | Qiu, W, Dong, A, Ren, H, Wu, H, Wasney, G, Vedadi, M, Lew, J, Kozieradski, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Plotnikov, A.N, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-25 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
4EB6
 
 | | Tubulin-Vinblastine: Stathmin-like complex | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ranaivoson, F.M, Gigant, B, Knossow, M. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structural plasticity of tubulin assembly probed by vinca-domain ligands.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TTS
 
 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus | | Descriptor: | Beta-galactosidase, ZINC ION | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
3TUC
 
 | | Crystal structure of SYK kinase domain with 1-benzyl-N-(5-(6,7-dimethoxyquinolin-4-yloxy)pyridin-2-yl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | Descriptor: | 1-benzyl-N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-2-oxo-1,2-dihydropyridine-3-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
3GQS
 
 | | Crystal structure of the FHA domain of CT664 protein from Chlamydia trachomatis | | Descriptor: | Adenylate cyclase-like protein, PHOSPHATE ION | | Authors: | Majorek, K.A, Cymborowski, M, Chruszcz, M, Evdokimova, E, Egorova, O, Di Leo, R, Zimmerman, M.D, Savchenko, A, Joachimiak, A, Edwards, A.M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the FHA domain of CT664 protein from Chlamydia trachomatis
To be Published
|
|
2L8M
 
 | | Reduced and CO-bound cytochrome P450cam (CYP101A1) | | Descriptor: | CAMPHOR, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Pochapsky, T.C, Pochapsky, S.S, Dang, M, Asciutto, E, Madura, J. | | Deposit date: | 2011-01-19 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Experimentally Restrained Molecular Dynamics Simulations for Characterizing the Open States of Cytochrome P450(cam).
Biochemistry, 50, 2011
|
|
2YJD
 
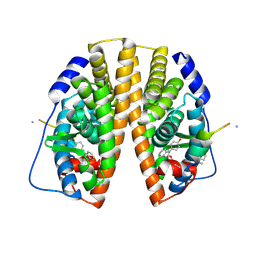 | | Stapled peptide bound to Estrogen Receptor Beta | | Descriptor: | 4-(2-PROPAN-2-YLOXYBENZIMIDAZOL-1-YL)PHENOL, ESTROGEN RECEPTOR BETA, STAPLED PEPTIDE | | Authors: | Phillips, C, Roberts, L.R, Schade, M, Bent, A, Davies, N.L, Moore, R, Pannifer, A.D, Brown, D.G, Pickford, A.R, Irving, S.L. | | Deposit date: | 2011-05-19 | | Release date: | 2011-08-03 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design and Structure of Stapled Peptides Binding to Estrogen Receptors.
J.Am.Chem.Soc., 133, 2011
|
|
3TV9
 
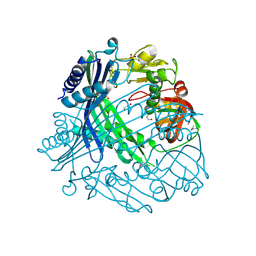 | | Crystal Structure of Putative Peptide Maturation Protein from Shigella flexneri | | Descriptor: | GLYCEROL, Putative peptide maturation protein, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-05 | | Last modified: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal Structure of Putative Peptide Maturation Protein from
To be Published
|
|
2YPD
 
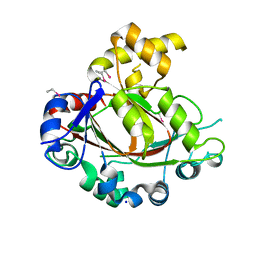 | | Crystal structure of the Jumonji domain of human Jumonji domain containing 1C protein | | Descriptor: | 2-(ETHYLMERCURI-THIO)-BENZOIC ACID, PROBABLE JMJC DOMAIN-CONTAINING HISTONE DEMETHYLATION PROT EIN 2C, SODIUM ION | | Authors: | Vollmar, M, Johansson, C, Krojer, T, Berridge, G, Burgess-Brown, N, Strain-Damerell, C, Froese, S, Williams, E, Goubin, S, Coutandin, D, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Jumonji Domain of Human Jumonji Domain Containing 1C Protein
To be Published
|
|
2YU8
 
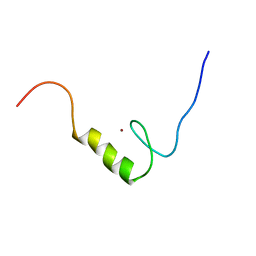 | | Solution structure of the C2H2 type zinc finger (region 648-680) of human Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 648-680) of human Zinc finger protein 347
To be Published
|
|
2LBH
 
 | | Solution Structure of the Dimeric Form of a Unliganded Bovine Neurophysin, Minimized Average Structure | | Descriptor: | Neurophysin 1 | | Authors: | Lee, H, Naik, M, Nguyen, T, Bracken, C, Breslow, E. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-04 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Dimerization-Induced Increase in Neurophysin-Hormone Affinity: Interplay of Inter-Domain and Inter-Subunit Interactions
To be Published
|
|
3TW2
 
 | | High resolution structure of human histidine triad nucleotide-binding protein 1 (hHINT1)/AMP complex in a monoclinic space group | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Wlodarczyk, A, Ozga, M, Krakowiak, A, Nawrot, B. | | Deposit date: | 2011-09-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A new crystal form of human histidine triad nucleotide-binding protein 1 (hHINT1) in complex with adenosine 5'-monophosphate at 1.38 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2YLS
 
 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: REDUCED ENZYME BOUND TO NADP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHENYLACETONE MONOOXYGENASE | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
2YVY
 
 | | Crystal structure of magnesium transporter MgtE cytosolic domain, Mg2+ bound form | | Descriptor: | MAGNESIUM ION, Mg2+ transporter MgtE | | Authors: | Hattori, M, Tanaka, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the MgtE Mg(2+) transporter
Nature, 448, 2007
|
|
2L3N
 
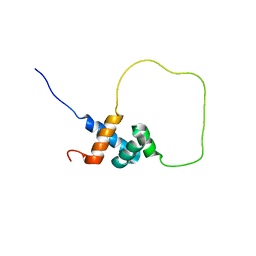 | | Solution structure of Rap1-Taz1 fusion protein | | Descriptor: | DNA-binding protein rap1,Telomere length regulator taz1 | | Authors: | Zhou, Z.R, Wang, F, Chen, Y, Lei, M, Hu, H. | | Deposit date: | 2010-09-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2L41
 
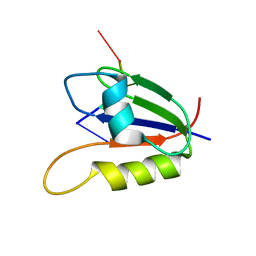 | | Nab3 RRM - UCUU complex | | Descriptor: | RNA (5'-R(P*UP*CP*UP*U)-3'), RRM domain from Nuclear polyadenylated RNA-binding protein 3 | | Authors: | Stefl, R, Pergoli, R, Hobor, F, Kubicek, K, Zimmermann, M, Pasulka, J, Hofr, C. | | Deposit date: | 2010-09-28 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein
J.Biol.Chem., 286, 2011
|
|
3TIC
 
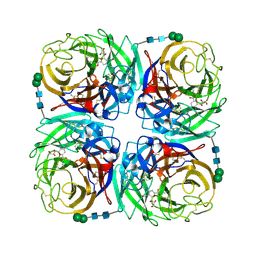 | | Crystal structure of 1957 pandemic H2N2 neuraminidase complexed with zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
2YXS
 
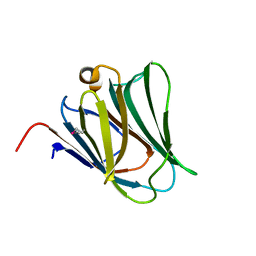 | | Crystal Structure of N-terminal domain of human galectin-8 with D-lactose | | Descriptor: | Galectin-8 variant, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of N-terminal domain of human galectin-8 with D-lactose
To be Published
|
|
3TIA
 
 | | Crystal structure of 1957 pandemic H2N2 neuraminidase complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
2YPN
 
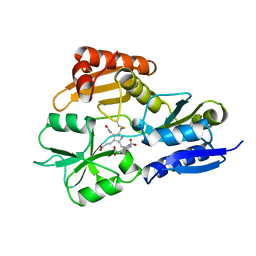 | | Hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, PROTEIN (HYDROXYMETHYLBILANE SYNTHASE) | | Authors: | Nieh, Y.P, Raftery, J, Weisgerber, S, Habash, J, Schotte, F, Ursby, T, Wulff, M, Haedener, A, Campbell, J.W, Hao, Q, Helliwell, J.R. | | Deposit date: | 1999-01-11 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Accurate and highly complete synchrotron protein crystal Laue diffraction data using the ESRF CCD and the Daresbury Laue software
J.Synchrotron Radiat., 6, 1999
|
|
3GNU
 
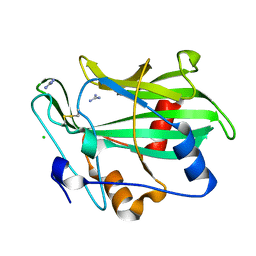 | | Toxin fold as basis for microbial attack and plant defense | | Descriptor: | 25 kDa protein elicitor, CHLORIDE ION, GUANIDINE | | Authors: | Ottmann, C, Luberacki, B, Kuefner, I, Koch, W, Brunner, F, Weyand, M, Mattinen, L, Pirhonen, M, Anderluh, G, Seitz, H.U, Nuernberger, T, Oecking, C. | | Deposit date: | 2009-03-18 | | Release date: | 2009-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A common toxin fold mediates microbial attack and plant defense
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3TKH
 
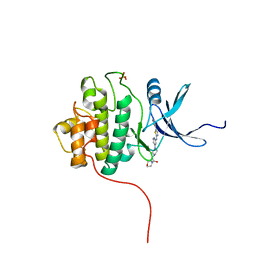 | | Crystal structure of Chk1 in complex with inhibitor S01 | | Descriptor: | 1-(morpholin-4-yl)-2-[4-(2-{[5-(pyridin-3-yl)-1,3-thiazol-2-yl]amino}pyridin-4-yl)piperazin-1-yl]ethanone, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Ikuta, M. | | Deposit date: | 2011-08-26 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Pyridyl aminothiazoles as potent inhibitors of Chk1 with slow dissociation rates.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
