4LWX
 
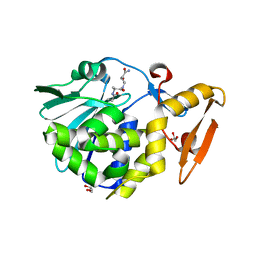 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica Balsamina with peptidoglycan fragment at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALANINE, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-29 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica Balsamina with peptidoglycan fragment at 1.78 A resolution
To be Published
|
|
3S5M
 
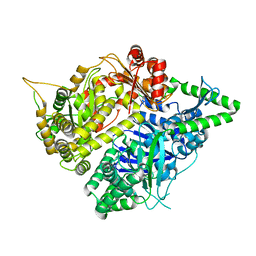 | | Crystal structures of falcilysin, a M16 metalloprotease from the malaria parasite Plasmodium falciparum | | Descriptor: | Falcilysin, MAGNESIUM ION, ZINC ION | | Authors: | Morgunova, E, Ponpuak, M, Istvan, E, Popov, A, Goldberg, D, Eneqvist, T. | | Deposit date: | 2011-05-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of falcilysin, a M16 metalloprotease from
the malaria parasite Plasmodium falciparum
To be Published
|
|
4LX8
 
 | |
1J9O
 
 | | SOLUTION STRUCTURE OF HUMAN LYMPHOTACTIN | | Descriptor: | LYMPHOTACTIN | | Authors: | Kuloglu, E.S, McCaslin, D.R, Kitabwalla, M, Pauza, C.D, Markley, J.L, Volkman, B.F. | | Deposit date: | 2001-05-28 | | Release date: | 2001-10-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Monomeric solution structure of the prototypical 'C' chemokine lymphotactin.
Biochemistry, 40, 2001
|
|
3GFH
 
 | |
4LXR
 
 | | Structure of the Toll - Spatzle complex, a molecular hub in Drosophila development and innate immunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stelter, M, Parthier, C, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2013-07-30 | | Release date: | 2014-04-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Toll-Spatzle complex, a molecular hub in Drosophila development and innate immunity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1JAF
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C' FROM RHODOCYCLUS GELATINOSUS AT 2.5 ANGSTOMS RESOLUTION | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Archer, M, Banci, L, Dikaya, E, Romao, M.J. | | Deposit date: | 1997-06-24 | | Release date: | 1998-01-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Cytochrome C' from Rhodocyclus Gelatinosus and Comparison with Other Cytochromes C'
J.Biol.Inorg.Chem., 2, 1997
|
|
3GGR
 
 | | Crystal Structure of the Human Rad9-Hus1-Rad1 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1 | | Authors: | Xu, M, Bai, L, Hang, H.Y, Jiang, T. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and functional implications of the human rad9-hus1-rad1 cell cycle checkpoint complex
J.Biol.Chem., 284, 2009
|
|
3GH5
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, beta-hexosaminidase | | Authors: | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-03-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
1JBH
 
 | | Solution structure of cellular retinol binding protein type-I in the ligand-free state | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN TYPE I | | Authors: | Franzoni, L, Luecke, C, Perez, C, Cavazzini, D, Rademacher, M, Ludwig, C, Spisni, A, Rossi, G.L, Rueterjans, H. | | Deposit date: | 2001-06-04 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of Apo- and holo-cellular retinol-binding protein in solution.
J.Biol.Chem., 277, 2002
|
|
1J9C
 
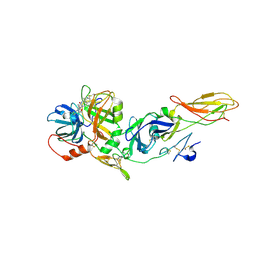 | | Crystal Structure of tissue factor-factor VIIa complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-phenylalaninamide, ... | | Authors: | Huang, M, Ruf, W, Edgington, T.S, Wilson, I.A. | | Deposit date: | 2001-05-24 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand Induced Conformational Transitions of Tissue Factor. Crystal Structure of the Tissue Factor:Factor VIIa Complex.
To be Published
|
|
1JCJ
 
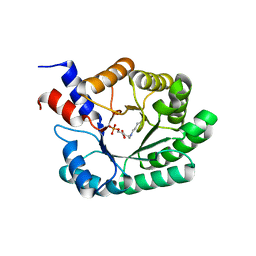 | | OBSERVATION OF COVALENT INTERMEDIATES IN AN ENZYME MECHANISM AT ATOMIC RESOLUTION | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, DEOXYRIBOSE-PHOSPHATE ALDOLASE | | Authors: | Heine, A, DeSantis, G, Luz, J.G, Mitchell, M, Wong, C.-H, Wilson, I.A. | | Deposit date: | 2001-06-09 | | Release date: | 2001-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
Science, 294, 2001
|
|
3G5L
 
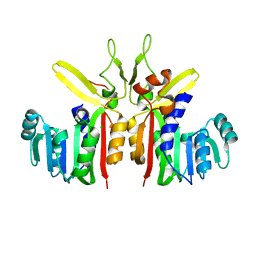 | | Crystal structure of putative S-adenosylmethionine dependent methyltransferase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Putative S-adenosylmethionine dependent methyltransferase | | Authors: | Patskovsky, Y, Sampathkumar, P, Gilmore, M, Miller, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of S-Adenosylmethionine Dependent Methyltransferase from Listeria Monocytogenes
To be Published
|
|
1JAE
 
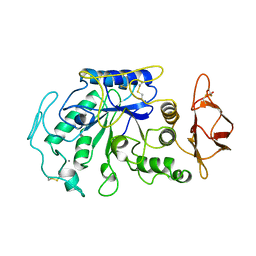 | | STRUCTURE OF TENEBRIO MOLITOR LARVAL ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Strobl, S, Maskos, K, Betz, M, Wiegand, G, Huber, R, Gomis-Rueth, F.X, Frank, G, Glockshuber, R. | | Deposit date: | 1997-09-30 | | Release date: | 1998-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of yellow meal worm alpha-amylase at 1.64 A resolution.
J.Mol.Biol., 278, 1998
|
|
3S95
 
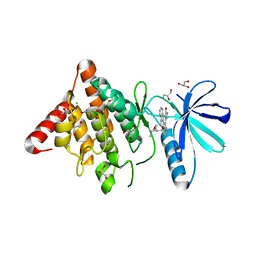 | | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Beltrami, A, Chaikuad, A, Daga, N, Elkins, J.M, Mahajan, P, Savitsky, P, Vollmar, M, Krojer, T, Muniz, J.R.C, Fedorov, O, Allerston, C.K, Yue, W.W, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine
To be Published
|
|
3GFZ
 
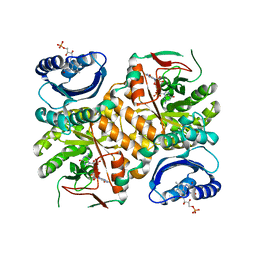 | | Klebsiella pneumoniae BlrP1 pH 6 manganese/cy-diGMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FLAVIN MONONUCLEOTIDE, Klebsiella pneumoniae BlrP1, ... | | Authors: | Barends, T, Hartmann, E, Griese, J, Beitlich, T, Kirienko, N, Ryjenkov, D, Reinstein, J, Shoeman, R, Gomelsky, M, Schlichting, I. | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase.
Nature, 459, 2009
|
|
4Q1J
 
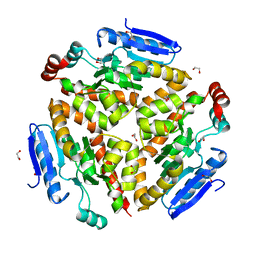 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 1,2-ETHANEDIOL, Polyketide biosynthesis enoyl-CoA isomerase PksI, SODIUM ION | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
4M3F
 
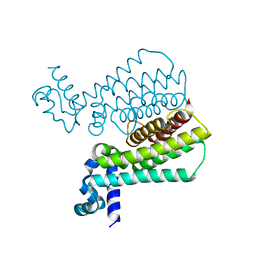 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | HTH-type transcriptional regulator EthR, N-(3-methylbutyl)-4-(2-methyl-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4Q22
 
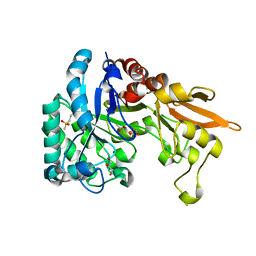 | | Crystal structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine at 1.93 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Madhuprakash, J, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2014-04-05 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine at 1.93 Angstrom resolution
To be Published
|
|
4Q2H
 
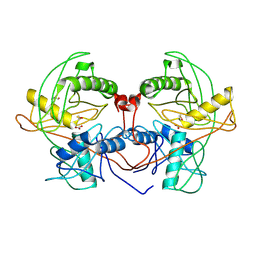 | | Crystal structure of probable proline racemase from agrobacterium radiobacter K84, TARGET EFI-506561, with bound carbonate | | Descriptor: | BICARBONATE ION, GLYCEROL, Proline racemase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Proline Racemase Arad_0731 from Agrobacterium Radiobacter, Target Efi-506561
To be Published
|
|
3SBB
 
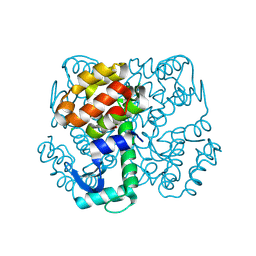 | | Disulphide-mediated Tetramer of T4 Lysozyme R76C/R80C by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, Lysozyme | | Authors: | Laganowsky, A, Soriaga, A.B, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (1.434 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3ZLQ
 
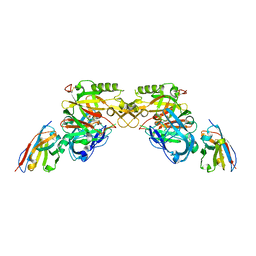 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 5-Ethoxy-pyridine-2-carboxylic acid [3-((R)-2-amino-5,5-difluoro-4-methyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide, BETA-SECRETASE 2, XA4813 | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-01 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Beta-Secretase (Bace1) Inhibitors with High in Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer'S Disease.
J.Med.Chem., 56, 2013
|
|
1J9H
 
 | | Crystal Structure of an RNA Duplex with Uridine Bulges | | Descriptor: | 5'-R(*GP*UP*GP*UP*CP*GP*(CBR)P*AP*C)-3', CALCIUM ION | | Authors: | Xiong, Y, Deng, J, Sudarsanakumar, C, Sundaralingam, M. | | Deposit date: | 2001-05-25 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an RNA duplex r(gugucgcac)(2) with uridine bulges.
J.Mol.Biol., 313, 2001
|
|
1J9W
 
 | | Solution Structure of the CAI Michigan 1 Variant | | Descriptor: | 1,2-ETHANEDIOL, CARBONIC ANHYDRASE I, ZINC ION | | Authors: | Briganti, F, Ferraroni, M, Chedwiggen, W.R, Scozzafava, A, Supuran, C.T, Tilli, S. | | Deposit date: | 2001-05-29 | | Release date: | 2001-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a zinc-activated variant of human carbonic anhydrase I, CA I Michigan 1: evidence for a second zinc binding site involving arginine coordination.
Biochemistry, 41, 2002
|
|
4Q39
 
 | | PylD in complex with pyrrolysine and NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Quitterer, F, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2014-04-11 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Formation of Pyrroline and Tetrahydropyridine Rings in Amino Acids Catalyzed by Pyrrolysine Synthase (PylD).
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
