4UT2
 
 | | X-ray structure of the human PP1 gamma catalytic subunit treated with ascorbate | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Kopec, J, Zeh Silva, M, Fotinou, C, Steiner, R.A. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeted Redox Inhibition of Protein Phosphatase 1 by Nox4 Regulates Eif2Alpha-Mediated Stress Signaling.
Embo J., 35, 2016
|
|
4WBL
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation F235A | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION, GLYCEROL | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
5WS5
 
 | | Native XFEL structure of photosystem II (preflash dark dataset) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
5J5Y
 
 | | Translation initiation factor 4E in complex with m2(7,2'O)GppCCl2ppG mRNA 5' cap analog | | Descriptor: | 2-amino-9-{5-O-[(R)-{[(S)-{dichloro[(R)-hydroxy(phosphonooxy)phosphoryl]methyl}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2-O-methyl-beta-D-ribofuranosyl}-7-methyl-9H-purin-7-ium-6-olate, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Rydzik, A.M, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-04-04 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | mRNA cap analogues substituted in the tetraphosphate chain with CX2: identification of O-to-CCl2 as the first bridging modification that confers resistance to decapping without impairing translation.
Nucleic Acids Res., 45, 2017
|
|
3B7K
 
 | | Human Acyl-coenzyme A thioesterase 12 | | Descriptor: | Acyl-coenzyme A thioesterase 12, COENZYME A | | Authors: | Lehtio, L, Busam, R.D, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human Acyl-coenzyme A thioesterase 12.
To be Published
|
|
1QH0
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 76 MUTATED BY ASP AND LEU 78 MUTATED BY ASP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
4UQG
 
 | | A new bio-isosteric base pair based on reversible bonding | | Descriptor: | 5'-D(*AP*GP*GP*GP*A SAYP*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*C T0TP*TP*CP*CP*CP*TP)-3', DNA POLYMERASE, ... | | Authors: | Tomas-Gamasa, M, Serdjukov, S, Su, M, Mueller, M, Carell, T. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | "Post-it" type connected DNA created with a reversible covalent cross-link.
Angew. Chem. Int. Ed. Engl., 54, 2015
|
|
4UV9
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Ethyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS,10aS)-4a-[(1S,3E)-3-imino-1-phenylpentyl]-7,8-dimethyl-2,4-dioxo-1,3,4,4a,5,10a-hexahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4V8V
 
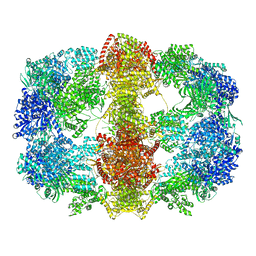 | | Structure and conformational variability of the Mycobacterium tuberculosis fatty acid synthase multienzyme complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, TYPE-I FATTY ACID SYNTHASE | | Authors: | Ciccarelli, L, Connell, S.R, Enderle, M, Mills, D.J, Vonck, J, Grininger, M. | | Deposit date: | 2013-04-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure and Conformational Variability of the Mycobacterium Tuberculosis Fatty Acid Synthase Multienzyme Complex.
Structure, 21, 2013
|
|
4WDF
 
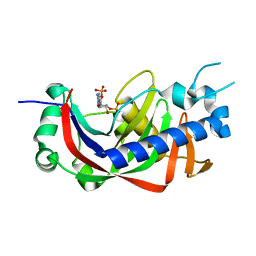 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation V321A, complexed with 2',5'-ADP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-5'-DIPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4UVB
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1S,2R) | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,7aS)-1-amino-1,10,11-trimethyl-4,6-dioxo-3-phenyl-2,3,5,6,7,7a-hexahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4WDH
 
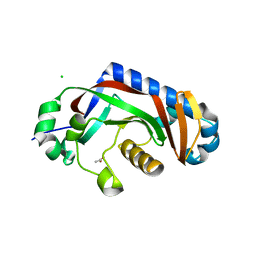 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation Y168A | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ACETIC ACID, CHLORIDE ION | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WDG
 
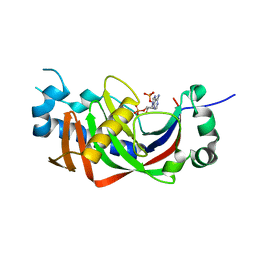 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation V321A, complexed with 2',5'-ADP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-5'-DIPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WDD
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation T232A, complexed with citrate | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
1QUW
 
 | | SOLUTION STRUCTURE OF THE THIOREDOXIN FROM BACILLUS ACIDOCALDARIUS | | Descriptor: | THIOREDOXIN | | Authors: | Nicastro, G, de Chiara, C, Pedone, E, Tato, M, Rossi, M. | | Deposit date: | 1999-07-02 | | Release date: | 2000-01-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a novel thioredoxin from Bacillus acidocaldarius possible determinants of protein stability.
Eur.J.Biochem., 267, 2000
|
|
4UZE
 
 | |
5WS6
 
 | | Native XFEL structure of Photosystem II (preflash two-flash dataset | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
1QJ4
 
 | |
2AMX
 
 | | Crystal structure of Plasmodium Yoelii Adenosine deaminase (PY02076) | | Descriptor: | COBALT (II) ION, UNKNOWN ATOM OR ION, adenosine deaminase | | Authors: | Dong, A, Vedadi, M, Wasney, G, Zhao, Y, Lew, J, Alam, Z, Melone, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-10 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1KCF
 
 | | Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2 | | Descriptor: | HYPOTHETICAL 30.2 KD PROTEIN C25G10.02 IN CHROMOSOME I, SULFATE ION | | Authors: | Ceschini, S, Keeley, A, McAlister, M.S.B, Oram, M, Phelan, J, Pearl, L.H, Tsaneva, I.R, Barrett, T.E. | | Deposit date: | 2001-11-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.
EMBO J., 20, 2001
|
|
4V12
 
 | | Crystal structure of the MSMEG_6754 dehydratase from Mycobacterium smegmatis | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAOC LIKE DOMAIN PROTEIN | | Authors: | Blaise, M. | | Deposit date: | 2014-09-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A New Dehydratase Conferring Innate Resistance to Thiacetazone and Intra-Amoebal Survival of Mycobacterium Smegmatis.
Mol.Microbiol., 96, 2015
|
|
5J4K
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with 1-Indane-6-carboxylic acid | | Descriptor: | 2,3-dihydro-1H-indene-2-carboxylic acid, CALCIUM ION, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
2A4A
 
 | | Deoxyribose-phosphate aldolase from P. yoelii | | Descriptor: | deoxyribose-phosphate aldolase | | Authors: | Walker, J.R, Amani, M, Lew, J, Wiegelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Hui, R, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-28 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
6ZH7
 
 | | Crystal structure of fatty acid photodecarboxylase in the dark state determined by serial femtosecond crystallography at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Weik, M, Schlichting, I, Barends, T.R.M, Colletier, J.P. | | Deposit date: | 2020-06-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
