6IAK
 
 | | The crystal structure of the chicken CREB3 bZIP | | Descriptor: | Uncharacterized protein | | Authors: | Sabaratnam, K, Renner, M. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-11 | | Last modified: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Insights from the crystal structure of the chicken CREB3 bZIP suggest that members of the CREB3 subfamily transcription factors may be activated in response to oxidative stress.
Protein Sci., 28, 2019
|
|
4O41
 
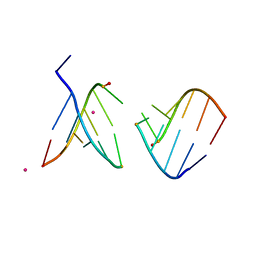 | | Amide linked RNA | | Descriptor: | AMIDE LINKED RNA, STRONTIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Amides are excellent mimics of phosphate internucleoside linkages and are well tolerated in short interfering RNAs.
Nucleic Acids Res., 42, 2014
|
|
3OSY
 
 | |
1HIK
 
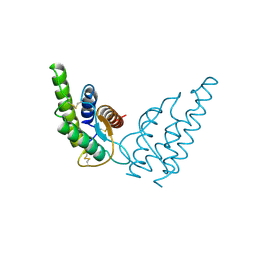 | | INTERLEUKIN-4 (WILD-TYPE) | | Descriptor: | INTERLEUKIN-4 | | Authors: | Mueller, T, Buehner, M. | | Deposit date: | 1995-06-01 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human interleukin-4 and variant R88Q: phasing X-ray diffraction data by molecular replacement using X-ray and nuclear magnetic resonance models.
J.Mol.Biol., 247, 1995
|
|
6IAR
 
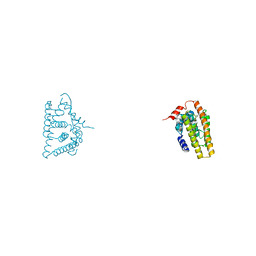 | | Tricyclic indazoles a novel class of selective estrogen receptor degrader antagonists | | Descriptor: | 3-[4-[(6~{R})-7-(2-methylpropyl)-3,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]phenyl]propanoic acid, Estrogen receptor | | Authors: | Scott, J.S, Bailey, A, Buttar, D, Carbajo, R.J, Curwen, J, Davies, R.D.M, Degorce, S.L, Donald, C, Gangl, E, Greenwood, R, Groombridge, S.D, Johnson, T, Lamont, S, Lawson, M, Lister, A, Morrow, C, Moss, T, Pink, J.H, Polanski, R. | | Deposit date: | 2018-11-27 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Tricyclic Indazoles-A Novel Class of Selective Estrogen Receptor Degrader Antagonists.
J.Med.Chem., 62, 2019
|
|
6RLR
 
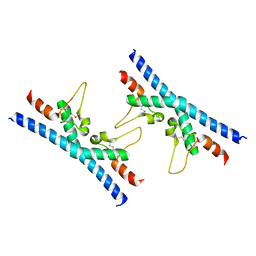 | | Crystal structure of CD9 large extracellular loop | | Descriptor: | CD9 antigen | | Authors: | Neviani, V, Kroon-Batenburg, L, Lutz, M, Pearce, N.M, Pos, W, Schotte, R, Spits, H, Gros, P. | | Deposit date: | 2019-05-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
3ETE
 
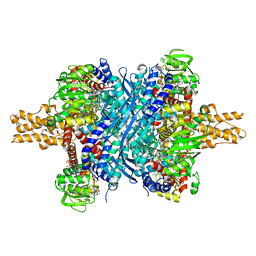 | | Crystal structure of bovine glutamate dehydrogenase complexed with hexachlorophene | | Descriptor: | 2,2'-methanediylbis(3,4,6-trichlorophenol), GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Li, M, Smith, T.J. | | Deposit date: | 2008-10-07 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel Inhibitors Complexed with Glutamate Dehydrogenase: ALLOSTERIC REGULATION BY CONTROL OF PROTEIN DYNAMICS
J.Biol.Chem., 284, 2009
|
|
6IBG
 
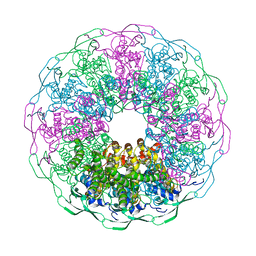 | | Bacteriophage G20c portal protein crystal structure for construct with intact N-terminus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Portal protein | | Authors: | Bayfield, O.W, Klimuk, E, Winkler, D.C, Hesketh, E.L, Chechik, M, Cheng, N, Dykeman, E.C, Minakhin, L, Ranson, N.A, Severinov, K, Steven, A.C, Antson, A.A. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1OMH
 
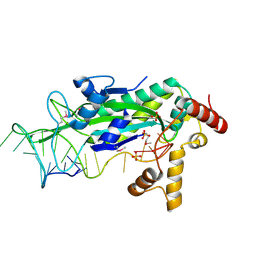 | | Conjugative Relaxase TrwC in complex with OriT Dna. Metal-free structure. | | Descriptor: | DNA OLIGONUCLEOTIDE, SULFATE ION, trwC protein | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2003-02-25 | | Release date: | 2003-11-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC.
Nat.Struct.Biol., 10, 2003
|
|
4NCV
 
 | | Foldon domain wild type N-conjugate | | Descriptor: | Fibritin | | Authors: | Graewert, M.A, Berthelmann, A, Lach, J, Groll, M, Eichler, J. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Versatile C(3)-symmetric scaffolds and their use for covalent stabilization of the foldon trimer.
Org.Biomol.Chem., 12, 2014
|
|
3E7V
 
 | | Crystal Structure of Human Haspin with a pyrazolo-pyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-aminophenyl)-N-(3-chlorophenyl)pyrazolo[1,5-a]pyrimidin-5-amine, NICKEL (II) ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wickstroem, M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Haspin with a pyrazolo-pyrimidine ligand
To be Published
|
|
3OVL
 
 | |
6IDF
 
 | | Cryo-EM structure of gamma secretase in complex with a Notch fragment | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Zhou, Q, Guo, X, Yan, C, Ke, M, Lei, J, Shi, Y. | | Deposit date: | 2018-09-09 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of Notch recognition by human gamma-secretase
Nature, 565, 2019
|
|
3E80
 
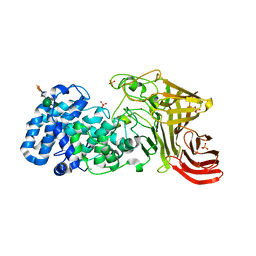 | |
6EHH
 
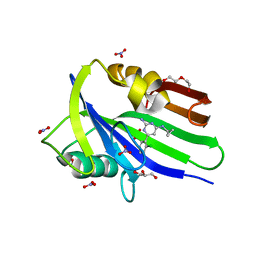 | | Crystal structure of mouse MTH1 mutant L116M with inhibitor TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gustafsson, R, Narwal, M, Jemth, A.-S, Almlof, I, Warpman Berglund, U, Helleday, T, Stenmark, P. | | Deposit date: | 2017-09-13 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Biochemistry, 57, 2018
|
|
4HUA
 
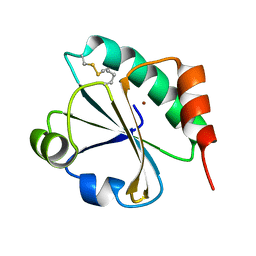 | | E. coli thioredoxin variant with (4R)-FluoroPro76 as single proline residue | | Descriptor: | COPPER (II) ION, Thioredoxin-1 | | Authors: | Scharer, M.A, Rubini, M, Capitani, G, Glockshuber, R. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | (4R)- and (4S)-Fluoroproline in the Conserved cis-Prolyl Peptide Bond of the Thioredoxin Fold: Tertiary Structure Context Dictates Ring Puckering.
Chembiochem, 14, 2013
|
|
3E2V
 
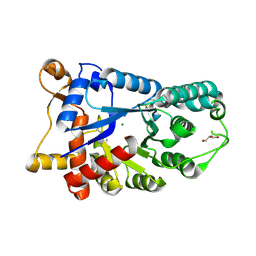 | | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae | | Descriptor: | 3'-5'-exonuclease, GLYCEROL, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-06 | | Release date: | 2008-08-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae
To be Published
|
|
4NM6
 
 | | Crystal structure of TET2-DNA complex | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*(5CM)P*GP*GP*TP*GP*GP*T)-3', FE (II) ION, Methylcytosine dioxygenase TET2, ... | | Authors: | Hu, L, Li, Z, Cheng, J, Rao, Q, Gong, W, Liu, M, Wang, P, Xu, Y. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation.
Cell(Cambridge,Mass.), 155, 2013
|
|
6EET
 
 | | Crystal structure of mouse Protocadherin-15 EC9-MAD12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Narui, Y, Sotomayor, M. | | Deposit date: | 2018-08-15 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural determinants of protocadherin-15 mechanics and function in hearing and balance perception.
Proc.Natl.Acad.Sci.USA, 2020
|
|
6RQ4
 
 | | Inhibitor of ERK2 | | Descriptor: | 6,6-dimethyl-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-5-(2-morpholin-4-ylethyl)thieno[2,3-c]pyrrol-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M. | | Deposit date: | 2019-05-15 | | Release date: | 2019-12-04 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Dual-Mechanism ERK1/2 Inhibitors Exploit a Distinct Binding Mode to Block Phosphorylation and Nuclear Accumulation of ERK1/2.
Mol.Cancer Ther., 19, 2020
|
|
1IL1
 
 | | Crystal structure of G3-519, an anti-HIV monoclonal antibody | | Descriptor: | monoclonal antibody G3-519 (heavy chain), monoclonal antibody G3-519 (light chain) | | Authors: | Berry, M.B, Johnson, K.A, Radding, W, Fung, M, Liou, R, Phillips Jr, G.N. | | Deposit date: | 2001-05-07 | | Release date: | 2001-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an anti-HIV monoclonal Fab antibody fragment specific to a gp120 C-4 region peptide.
Proteins, 45, 2001
|
|
1ILF
 
 | |
6S6D
 
 | | Crystal structure of RagA-Q66L-GTP/RagC-S75N-GDP GTPase heterodimer complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Anandapadamanaban, M, Masson, G.R, Perisic, O, Kaufman, J, Williams, R.L. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
6S6U
 
 | |
4HVY
 
 | | A thermostable variant of human NUDT18 NUDIX domain obtained by Hot Colony Filtration | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nucleoside diphosphate-linked moiety X motif 18, ... | | Authors: | Asial, I, Cheng, Y.X, Engman, H, Wu, B, Dollhopf, M, Nordlund, P, Cornvik, T. | | Deposit date: | 2012-11-07 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Engineering protein thermostability using a generic activity-independent biophysical screen inside the cell.
Nat Commun, 4, 2013
|
|
