5FTX
 
 | | Structure of surface layer protein SbsC, domains 4-9 | | Descriptor: | CALCIUM ION, SURFACE LAYER PROTEIN, ZINC ION | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2016-01-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of Surface Layer Protein Sbsc
To be Published
|
|
5CKY
 
 | | Crystal Structure of the MTERF1 R162A substitution bound to the termination sequence. | | Descriptor: | 5' -D (*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3', 5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3', Transcription termination factor 1, ... | | Authors: | Byrnes, J, Hauser, K, Norona, L, Mejia, E, Simmerling, C, Garcia-Diaz, M. | | Deposit date: | 2015-07-15 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
J.Mol.Biol., 428, 2016
|
|
4A24
 
 | | Structural and functional analysis of the DEAF-1 and BS69 MYND domains | | Descriptor: | DEFORMED EPIDERMAL AUTOREGULATORY FACTOR 1 HOMOLOG, ZINC ION | | Authors: | Kateb, F, Perrin, H, Tripsianes, K, Zou, P, Spadaccini, R, Bottomley, M, Bepperling, A, Ansieau, S, Sattler, M. | | Deposit date: | 2011-09-22 | | Release date: | 2012-11-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Analysis of the Deaf-1 and Bs69 Mynd Domains.
Plos One, 8, 2013
|
|
5FWS
 
 | | Wnt modulator Kremen crystal form I at 1.90A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
6B4T
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 4-methylpyridin-2-ol | | Descriptor: | 4-methylpyridin-2-ol, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 4-methylpyridin-2-ol
To Be Published
|
|
5O74
 
 | | Crystal structure of human Rab1b covalently bound to the GEF domain of DrrA/SidM from Legionella pneumophila in the presence of GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Multifunctional virulence effector protein DrrA, Ras-related protein Rab-1B | | Authors: | Cigler, M, Mueller, T, Horn-Ghetko, D, von Wrisberg, M.K, Fottner, M, Goody, R.S, Itzen, A, Mueller, M.P, Lang, K. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Proximity-Triggered Covalent Stabilization of Low-Affinity Protein Complexes In Vitro and In Vivo.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5FQL
 
 | | Insights into Hunter syndrome from the structure of iduronate-2- sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Demydchuk, M, Hill, C.H, Zhou, A, Bunkoczi, G, Stein, P.E, Marchesan, D, Deane, J.E, Read, R.J. | | Deposit date: | 2015-12-11 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into Hunter syndrome from the structure of iduronate-2-sulfatase.
Nat Commun, 8, 2017
|
|
6B63
 
 | | IMPase (AF2372) with 25 mM Asp | | Descriptor: | (2R)-2,3-dihydroxypropyl (2S)-2,3-dihydroxypropyl hydrogen phosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fructose-1,6-bisphosphatase/inositol-1-monophosphatase, ... | | Authors: | Goldstein, R.I, Roberts, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Osmolyte binding capacity of a dual action IMPase/FBPase (AF2372)
To Be Published
|
|
4IN9
 
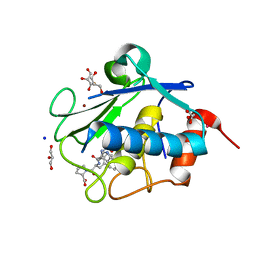 | | Structure of karilysin MMP-like catalytic domain in complex with inhibitory tetrapeptide SWFP | | Descriptor: | GLYCEROL, Karilysin protease, POTASSIUM ION, ... | | Authors: | Guevara, T, Ksiazek, M, Skottrup, P.D, Cerda-Costa, N, Trillo-Muyo, S, de Diego, I, Riise, E, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2013-01-04 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the catalytic domain of the Tannerella forsythia matrix metallopeptidase karilysin in complex with a tetrapeptidic inhibitor.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6V71
 
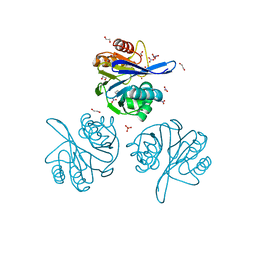 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site
To Be Published
|
|
5FYW
 
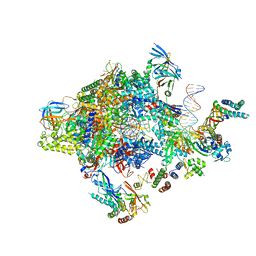 | | Transcription initiation complex structures elucidate DNA opening (OC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
6V2J
 
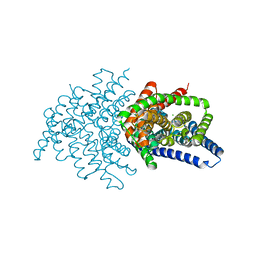 | | Crystal structure of ClC-ec1 triple mutant (E113Q, E148Q, E203Q) | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Maduke, M, Mathews, I.I, Chavan, T.S. | | Deposit date: | 2019-11-24 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A CLC-ec1 mutant reveals global conformational change and suggests a unifying mechanism for the CLC Cl - /H + transport cycle.
Elife, 9, 2020
|
|
4IOU
 
 | |
6B82
 
 | | Zebra Fish CYP-450 17A1 Mutant Abiraterone Complex | | Descriptor: | ACETATE ION, Abiraterone, CHLORIDE ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-10-05 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Inherent steroid 17 alpha ,20-lyase activity in defunct cytochrome P450 17A enzymes.
J. Biol. Chem., 293, 2018
|
|
6V7S
 
 | | Crystal structure of K37-acetylated SUMO1 in complex with phosphorylated PIAS-SIM2 | | Descriptor: | Protein PIAS, Small ubiquitin-related modifier 1 | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure, 28, 2020
|
|
7RHM
 
 | |
6V8L
 
 | | Crystal structure of Ara h 8.0201 | | Descriptor: | Ara h 8 allergen isoform, SULFATE ION, icosanoic acid | | Authors: | Pote, S, Offermann, L.R, Hurlburt, B.K, McBride, J.K, Chruszcz, M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Ara h 8.0201
To Be Published
|
|
4ACG
 
 | | GSK3b in complex with inhibitor | | Descriptor: | 2-AMINO-5-{4-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-N-[4-(PYRROLIDIN-1-YLMETHYL)PYRIDIN-3-YL]PYRIDINE-3-CARBOXAMIDE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Xue, Y, Ormo, M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
6VCX
 
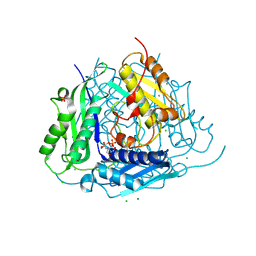 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 1 (AtMAT1) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
7RAO
 
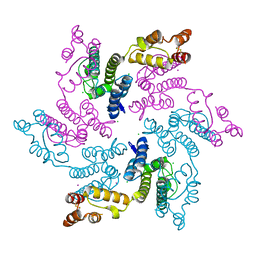 | |
6BAR
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA (Q5SIX3_THET8) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Rod shape determining protein RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
6V3U
 
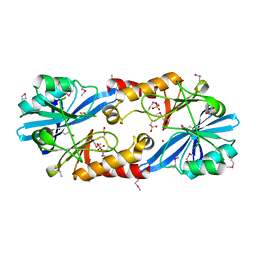 | | Crystal Structure of the NDM_FIM-1 like Metallo-beta-Lactamase from Erythrobacter litoralis in the Mono-Zinc Form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase II, ISOPROPYL ALCOHOL, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-26 | | Release date: | 2020-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the NDM_FIM-1 like Metallo-beta-Lactamase from Erythrobacter litoralis in the Mono-Zinc Form
To Be Published
|
|
8B7I
 
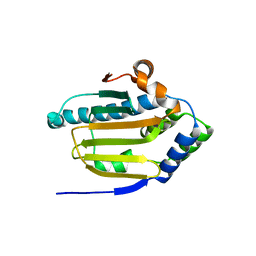 | | Human HSP90 alpha ATP Binding Domain, ATP-lid open conformation, R60A | | Descriptor: | HSP90AA1 protein | | Authors: | Rioual, E, Henot, F, Favier, A, Macek, P, Crublet, E, Josso, P, Brutscher, B, Frech, M, Gans, P, Loison, C, Boisbouvier, J. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Visualizing the transiently populated closed-state of human HSP90 ATP binding domain.
Nat Commun, 13, 2022
|
|
5GS4
 
 | | Crystal structure of estrogen receptor alpha in complex with a stabilized peptide antagonist | | Descriptor: | ARG-IAS-ILE-LEU-DNP-ARG-LEU-LEU-GLN, ESTRADIOL, Estrogen receptor, ... | | Authors: | Xie, M, Wang, T, Li, Z.-G. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-30 | | Last modified: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Inhibition of ER alpha-Coactivator Interaction by High-Affinity N-Terminus Isoaspartic Acid Tethered Helical Peptides
J. Med. Chem., 60, 2017
|
|
6B64
 
 | | IMPase (AF2372) with 25 mM Asp | | Descriptor: | ASPARTIC ACID, Fructose-1,6-bisphosphatase/inositol-1-monophosphatase, MAGNESIUM ION, ... | | Authors: | Goldstein, R.I, Roberts, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Osmolyte binding capacity of a dual action IMPase/FBPase (AF2372)
To Be Published
|
|
