6WNR
 
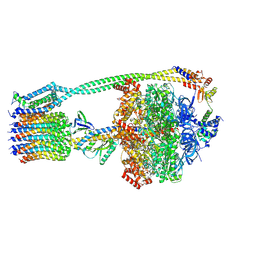 | | E. coli ATP synthase State 3b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6EZE
 
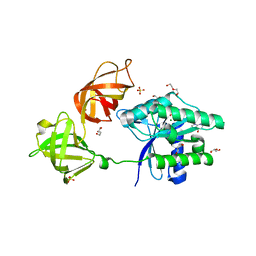 | | The open conformation of E.coli Elongation Factor Tu in complex with GDPNP. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongation factor Tu 2, GLYCEROL, ... | | Authors: | Johansen, J.S, Blaise, M, Thirup, S.S. | | Deposit date: | 2017-11-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | E. coli elongation factor Tu bound to a GTP analogue displays an open conformation equivalent to the GDP-bound form.
Nucleic Acids Res., 46, 2018
|
|
5TTX
 
 | |
5JEI
 
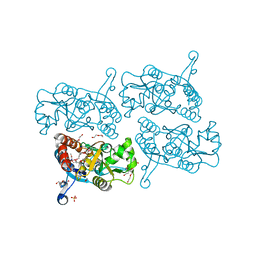 | | Crystal structure of the GluA2 LBD in complex with FW | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, ... | | Authors: | Eibl, C, Salazar, H, Chebli, M, Plested, A.J.R. | | Deposit date: | 2016-04-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.229 Å) | | Cite: | Mechanism of partial agonism in AMPA-type glutamate receptors.
Nat Commun, 8, 2017
|
|
6ETM
 
 | |
6WPG
 
 | | Structural Basis of Salicylic Acid Perception by Arabidopsis NPR Proteins | | Descriptor: | 2-HYDROXYBENZOIC ACID, Regulatory protein NPR4 | | Authors: | Wang, W, Withers, J, Li, H, Zwack, P.J, Rusnac, D.V, Shi, H, Liu, L, Yan, S, Hinds, T.R, Guttman, M, Dong, X, Zheng, N. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.283 Å) | | Cite: | Structural basis of salicylic acid perception by Arabidopsis NPR proteins.
Nature, 586, 2020
|
|
6BAR
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA (Q5SIX3_THET8) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Rod shape determining protein RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
5J3V
 
 | |
6EUV
 
 | |
6EZT
 
 | | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase D437A inactive mutant from Vibrio harveyi | | Descriptor: | Beta-N-acetylglucosaminidase Nag2, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Porfetye, A.T, Meekrathok, P, Burger, M, Vetter, I.R, Suginta, W. | | Deposit date: | 2017-11-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi
To Be Published
|
|
5U3R
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 2 | | Descriptor: | 6-[2-({[4-(furan-2-yl)benzene-1-carbonyl](propan-2-yl)amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6WZ5
 
 | |
5U43
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 12 | | Descriptor: | 6-(2-{[cyclopropyl(4'-methoxy[1,1'-biphenyl]-4-carbonyl)amino]methyl}phenoxy)hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6EWG
 
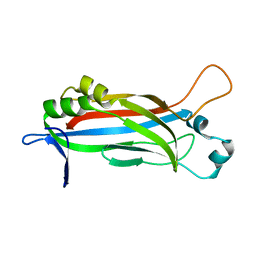 | |
6B04
 
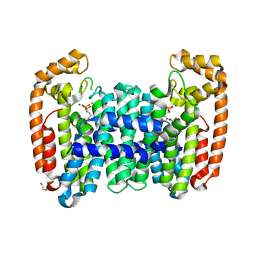 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with [2-(1-methylpyridin-2-yl)-1-phosphono-ethyl]phosphonic acid (inhibitor 1b) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,2-diphosphonoethyl)-1-methylpyridin-1-ium, Farnesyl diphosphate synthase, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
5J5F
 
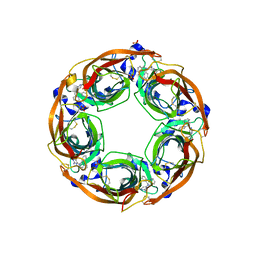 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with N4,N4-bis[(pyridin-2-yl)methyl]-6-(thiophen-3-yl)pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kaczanowska, K, Camacho Hernandez, G.A, Harel, M, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
5JG4
 
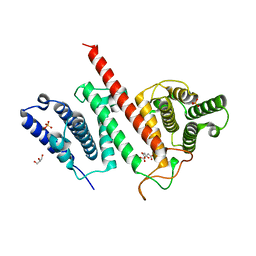 | | Structure of the effector protein LpiR1 (Lpg0634) from Legionella pneumophila | | Descriptor: | CITRATE ANION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Beyrakhova, K, van Straaten, K, Cygler, M. | | Deposit date: | 2016-04-19 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Investigations of the Effector Protein LpiR1 from Legionella pneumophila.
J.Biol.Chem., 291, 2016
|
|
6EX9
 
 | |
5TZQ
 
 | | Crystal Structure of FPV039:Bmf BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-like protein FPV039, Bcl-2-modifying factor | | Authors: | Anasir, M.I, Kvansakul, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039.
J. Biol. Chem., 292, 2017
|
|
6B2L
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with piperidin-2-Imine | | Descriptor: | DIMETHYL SULFOXIDE, PIPERIDIN-2-IMINE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with piperidin-2-Imine
To Be Published
|
|
6B3Y
 
 | | Crystal structure of the PH-like domain from DENND3 | | Descriptor: | DENN domain-containing protein 3 | | Authors: | Kozlov, G, Xu, J, Menade, M, Beaugrand, M, Pan, T, McPherson, P.S, Gehring, K. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | A PH-like domain of the Rab12 guanine nucleotide exchange factor DENND3 binds actin and is required for autophagy.
J. Biol. Chem., 293, 2018
|
|
7GAT
 
 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
6F12
 
 | | GLIC mutant E181A | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
6X0V
 
 | | Structure of MZT2/GCP-NHD and CDK5Rap2 at position 13 of the gamma-TuRC | | Descriptor: | Centrosome protein Cep215, Gamma-tubulin complex component 2, Mitotic-spindle organizing protein 2A | | Authors: | Wieczorek, M, Huang, T.-L, Urnavicius, L, Hsia, K.-C, Kapoor, T.M. | | Deposit date: | 2020-05-17 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | MZT Proteins Form Multi-Faceted Structural Modules in the gamma-Tubulin Ring Complex.
Cell Rep, 31, 2020
|
|
5U41
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 16 | | Descriptor: | 6-[2-({benzyl[4-(thiophen-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, POTASSIUM ION, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
