1WI0
 
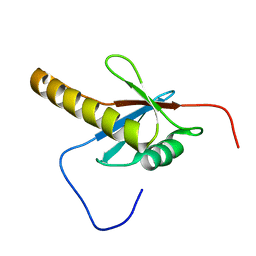 | | Solution structure of the PB1 domain of mouse mitogen activated protein kinase kinase 5 (MAP2K5) | | Descriptor: | Mitogen activated protein kinase kinase 5 | | Authors: | Yoneyama, M, Hayashi, F, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PB1 domain of mouse mitogen activated protein kinase kinase 5 (MAP2K5)
To be Published
|
|
7SFB
 
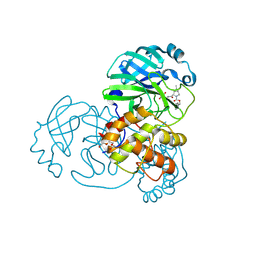 | |
4ZC3
 
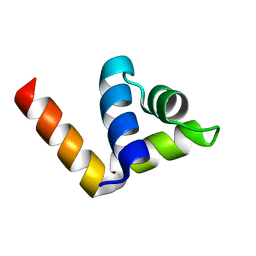 | |
7SFI
 
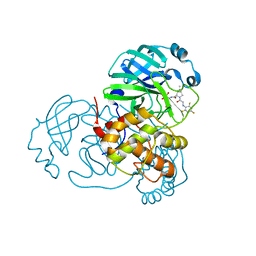 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML104 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[N-(2,4,6-trifluorophenyl)glycyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-03 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
7SB6
 
 | | Crystal Structure of Ancestral Mammalian Cadherin-23 EC1-2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin 23, ... | | Authors: | Nisler, C.R, Narui, Y, Sotomayor, M. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.588 Å) | | Cite: | Interpreting the Evolutionary Echoes of a Protein Complex Essential for Inner-Ear Mechanosensation.
Mol.Biol.Evol., 40, 2023
|
|
6U7N
 
 | | Crystal structure of neurotrimin (NTM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Neurotrimin, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G, Rush, S. | | Deposit date: | 2019-09-03 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
7SH4
 
 | | CD1a-phosphatidylglycerol binary structure | | Descriptor: | (21R,24R,27S)-24,27,28-trihydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaoctacosan-21-yl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wegrecki, M, Rossjohn, J. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylococcal phosphatidylglycerol antigens activate human T cells via CD1a.
Nat.Immunol., 24, 2023
|
|
6OEP
 
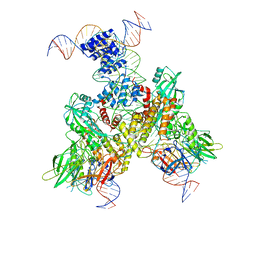 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OF4
 
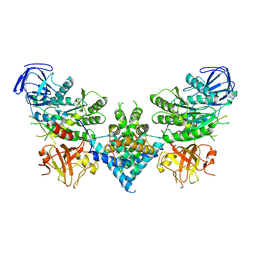 | | Precursor ribosomal RNA processing complex, apo-state. | | Descriptor: | CLP1_P domain-containing protein, Ribonuclease | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OGK
 
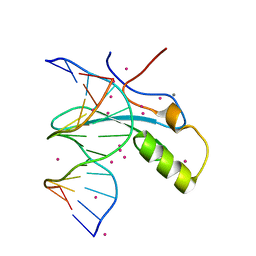 | | MeCP2 MBD in complex with DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
6LK9
 
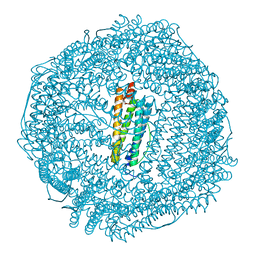 | | Coho salmon ferritin | | Descriptor: | Coho salmon ferritin, FE (III) ION | | Authors: | Wang, Z, Zang, J, Li, H, Tan, X, Du, M. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Coho salmon ferritin
To Be Published
|
|
1WKH
 
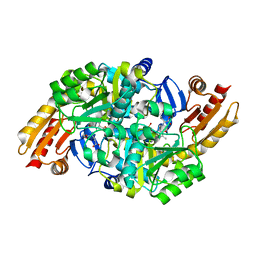 | | Acetylornithine aminotransferase from thermus thermophilus HB8 | | Descriptor: | 4-[(1,3-DICARBOXY-PROPYLAMINO)-METHYL]-3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDINIUM, Acetylornithine/acetyl-lysine aminotransferase | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Acetylornithine aminotransferase from thermus thermophilus HB8
To be Published
|
|
1X4L
 
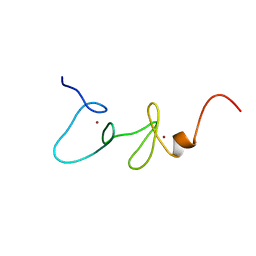 | | Solution structure of LIM domain in Four and a half LIM domains protein 2 | | Descriptor: | Skeletal muscle LIM-protein 3, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of LIM domain in Four and a half LIM domains protein 2
To be Published
|
|
7NBA
 
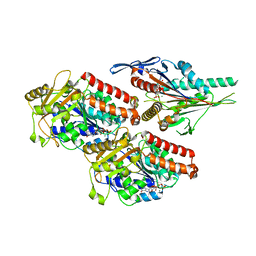 | | Plasmodium falciparum kinesin-5 motor domain bound to AMPPNP, complexed with 14 protofilament microtubule. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin motor domain-containing protein,Kinesin motor domain-containing protein, MAGNESIUM ION, ... | | Authors: | Cook, A.D, Roberts, A, Atherton, J, Tewari, R, Topf, M, Moores, C.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
J.Biol.Chem., 297, 2021
|
|
6MDO
 
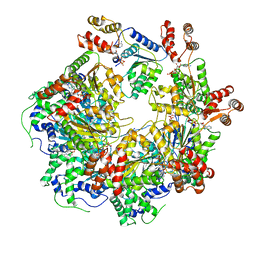 | | The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Synaptosomal-associated protein 25, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6UAH
 
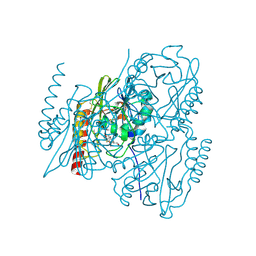 | | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem
To Be Published
|
|
6OUX
 
 | | Structure of SMUL_1544, a decarboxylase from Sulfurospirillum multivorans | | Descriptor: | Threonine phosphate decarboxylase-like enzyme | | Authors: | Wetterhorn, K.M, Rayment, I, Vecellio, A, Seeger, M, Keller, S, Schubert, T. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional analysis of an l-serine O-phosphate decarboxylase involved in norcobamide biosynthesis.
Febs Lett., 593, 2019
|
|
1X5P
 
 | | Solution structure of RRM domain in Parp14 | | Descriptor: | Negative elongation factor E | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in Parp14
To be Published
|
|
6EM9
 
 | | S.aureus ClpC resting state, asymmetric map | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6LMW
 
 | | Cryo-EM structure of the CALHM chimeric construct (8-mer) | | Descriptor: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
2LVZ
 
 | | Solution structure of a Eosinophil Cationic Protein-trisaccharide heparin mimetic complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-propan-2-yl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, Eosinophil cationic protein | | Authors: | Garcia Mayoral, M, Canales, A, Diaz, D, Lopez Prados, J, Moussaoui, M, de Paz, J, Angulo, J, Nieto, P, Jimenez Barbero, J, Boix, E, Bruix, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Insights into the glycosaminoglycan-mediated cytotoxic mechanism of eosinophil cationic protein revealed by NMR.
Acs Chem.Biol., 8, 2013
|
|
4OVR
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM XANTHOBACTER AUTOTROPHICUS PY2, TARGET EFI-510329, WITH BOUND BETA-D-GALACTURONATE | | Descriptor: | CHLORIDE ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
6UBH
 
 | | Structure of the MM7 Erbin PDZ variant in complex with a high-affinity peptide | | Descriptor: | Erbin, SODIUM ION, peptide | | Authors: | Singer, A.U, Teyra, J, McLaughlin, M, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-09-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comprehensive Assessment of the Relationship Between Site -2 Specificity and Helix alpha 2 in the Erbin PDZ Domain.
J.Mol.Biol., 433, 2021
|
|
7NB8
 
 | | Plasmodium falciparum kinesin-5 motor domain without nucleotide, complexed with 14 protofilament microtubule. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin-5, MAGNESIUM ION, ... | | Authors: | Cook, A.D, Roberts, A, Atherton, J, Tewari, R, Topf, M, Moores, C.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
J.Biol.Chem., 297, 2021
|
|
6OWF
 
 | | Structure of a synthetic beta-carboxysome shell, T=3 | | Descriptor: | Ethanolamine utilization protein EutN/carboxysome structural protein Ccml, Microcompartments protein | | Authors: | Sutter, M, Laughlin, T.G, Davies, K.M, Kerfeld, C.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a Syntheticbeta-Carboxysome Shell.
Plant Physiol., 181, 2019
|
|
