6SQ9
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 3-hydroxynaphthalene-2-carboxylic acid | | Descriptor: | 3-hydroxynaphthalene-2-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
7B73
 
 | | Insight into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Short-chain dehydrogenase/reductase SDR | | Authors: | Wessel, J, Petrillo, G, Estevez, M, Bosh, S, Seeger, M, Dijkman, W.P, Hidalgo, A, Uson, I, Osuna, S, Schallmey, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2.
Febs J., 288, 2021
|
|
5GVO
 
 | | Solution NMR structure of a new lasso peptide sphaericin | | Descriptor: | Uncharacterized protein | | Authors: | Hemmi, H, Kodani, S, Inoue, Y, Suzuki, M, Dohra, H, Suzuki, T, Ohnishi-Kameyama, M. | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-05 | | Method: | SOLUTION NMR | | Cite: | Sphaericin, a Lasso Peptide from the Rare Actinomycete Planomonospora sphaerica
Eur.J.Org.Chem., 2017
|
|
5MUD
 
 | | Crystal structure of an amyloidogenic light chain dimer H6 | | Descriptor: | light chain dimer,IGL@ protein | | Authors: | Oberti, L, Bacarizo, J, Maritan, M, Rognoni, P, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5GXG
 
 | | High-resolution crystal structure of the electron transfer complex of cytochrome p450cam with putidaredoxin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kikui, Y, Hiruma, Y, Ubbink, M, Nojiri, M. | | Deposit date: | 2016-09-17 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of productive and futile encounters in an electron transfer protein complex
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1WU4
 
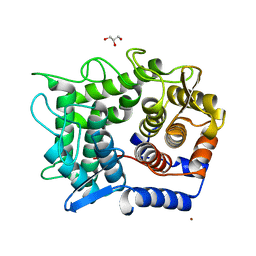 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase | | Descriptor: | GLYCEROL, NICKEL (II) ION, xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1WKY
 
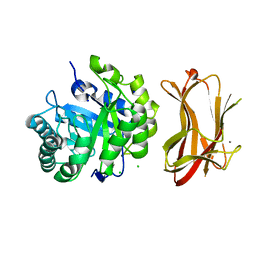 | | Crystal structure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Akita, M, Takeda, N, Hirasawa, K, Sakai, H, Kawamoto, M, Yamamoto, M, Grant, W.D, Hatada, Y, Ito, S, Horikoshi, K. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization and preliminary X-ray study of alkaline mannanase from an alkaliphilic Bacillus isolate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ZT3
 
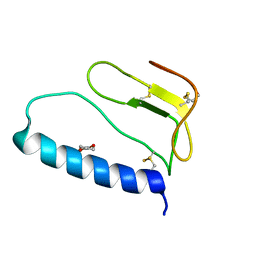 | | C-terminal domain of Insulin-like Growth Factor Binding Protein-1 isolated from human amniotic fluid | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Insulin-like growth factor binding protein 1 | | Authors: | Sala, A, Capaldi, S, Campagnoli, M, Faggion, B, Labo, S, Perduca, M, Romano, A, Carrizo, M.E, Valli, M, Visai, L, Minchiotti, L, Galliano, M, Monaco, H.L. | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Properties of the C-terminal Domain of Insulin-like Growth Factor-binding Protein-1 Isolated from Human Amniotic Fluid
J.Biol.Chem., 280, 2005
|
|
1ZS2
 
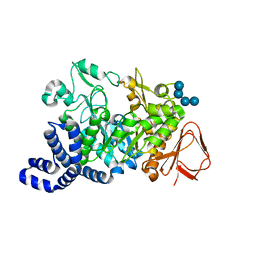 | | Amylosucrase Mutant E328Q in a ternary complex with sucrose and maltoheptaose | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, amylosucrase, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, van der Veen, B.A, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2005-05-23 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the Glu328Gln mutant of Neisseria polysaccharea amylosucrase in complex with sucrose and maltoheptaose
BIOCATAL.BIOTRANSFOR., 24, 2006
|
|
1WNX
 
 | | D136E mutant of Heme Oxygenase from Corynebacterium diphtheriae (HmuO) | | Descriptor: | Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Unno, M, Matsui, T, Ikeda-Saito, M. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of Distal Asp in Heme Oxygenase from Corynebacterium diphtheriae, HmuO: A WATER-DRIVEN OXYGEN ACTIVATION MECHANISM
J.Biol.Chem., 280, 2005
|
|
6F7U
 
 | | Molecular Mechanism of ATP versus GTP Selectivity of Adenylate Kinase | | Descriptor: | Adenylate kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Rogne, P, Rosselin, M, Grundstrom, C, Hedberg, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2017-12-12 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular mechanism of ATP versus GTP selectivity of adenylate kinase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1WT7
 
 | | Solution structure of BuTX-MTX: a butantoxin-maurotoxin chimera | | Descriptor: | BuTX-MTX | | Authors: | M'Barek, S, Chagot, B, Andreotti, N, Visan, V, Mansuelle, P, Grissmer, S, Marrakchi, M, El Ayeb, M, Sampieri, F, Darbon, H, Fajloun, Z, De Waard, M, Sabatier, J.-M. | | Deposit date: | 2004-11-16 | | Release date: | 2004-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Increasing the molecular contacts between maurotoxin and Kv1.2 channel augments ligand affinity.
Proteins, 60, 2005
|
|
1ZT5
 
 | | C-terminal domain of Insulin-like Growth Factor Binding Protein-1 isolated from human amniotic fluid complexed with Iron(II) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FE (II) ION, Insulin-like growth factor binding protein 1 | | Authors: | Sala, A, Capaldi, S, Campagnoli, M, Faggion, B, Labo, S, Perduca, M, Romano, A, Carrizo, M.E, Valli, M, Visai, L, Minchiotti, L, Galliano, M, Monaco, H.L. | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Structure and Properties of the C-terminal Domain of Insulin-like Growth Factor-binding Protein-1 Isolated from Human Amniotic Fluid
J.Biol.Chem., 280, 2005
|
|
6PFK
 
 | |
1WXU
 
 | | Solution structure of the SH3 domain of mouse peroxisomal biogenesis factor 13 | | Descriptor: | peroxisomal biogenesis factor 13 | | Authors: | Yoneyama, M, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-01 | | Release date: | 2005-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of mouse peroxisomal biogenesis factor 13
To be Published
|
|
8E1A
 
 | | Structure-based study to overcome cross-reactivity of novel androgen receptor inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(3-fluoro-2-methoxyphenyl)-1,3-thiazol-2-yl]morpholine, Androgen receptor | | Authors: | Lallous, N, Li, H, Radaeva, M, Dalal, K, Leblanc, E, Ban, F, Ciesielski, F, Chow, B, Morin, M, Singh, K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2022-08-10 | | Release date: | 2022-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Study to Overcome Cross-Reactivity of Novel Androgen Receptor Inhibitors.
Cells, 11, 2022
|
|
3LOF
 
 | | C-terminal domain of human heat shock 70kDa protein 1B. | | Descriptor: | Heat shock 70 kDa protein 1 | | Authors: | Osipiuk, J, Gu, M, Mihelic, M, Orton, K, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-03 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of C-terminal domain of human heat shock 70kDa protein 1B.
To be Published
|
|
6CG0
 
 | | Cryo-EM structure of mouse RAG1/2 HFC complex (3.17 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
3LQD
 
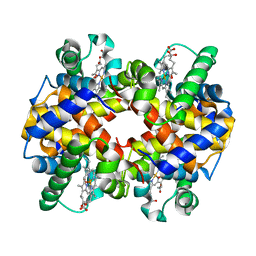 | | Crystal structure determination of Lepus europaeus 2.8 A resolution | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Thenmozhi, M, Sathya Moothy, Pon, Balasubramanian, M, Ponnuswamy, M.N. | | Deposit date: | 2010-02-09 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure determination of Lepus europaeus 2.8 A resolution
To be published
|
|
5N4K
 
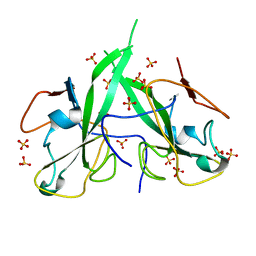 | | N-terminal domain of a human Coronavirus NL63 nucleocapsid protein | | Descriptor: | Nucleoprotein, SULFATE ION | | Authors: | Zdzalik, M, Szelazek, B, Kabala, W, Golik, P, Burmistrz, M, Florek, D, Kus, K, Pyrc, K, Dubin, G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
6F06
 
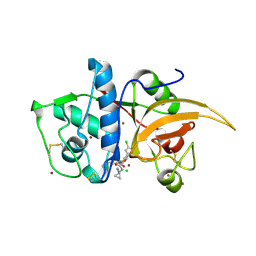 | | CATHEPSIN L IN COMPLEX WITH (3S,14E)-8-(azetidin-3-yl)-19-chloro-N-(1-cyanocyclopropyl)-5-oxo-12,17-dioxa-4-azatricyclo[16.2.2.06,11]docosa-1(21),6,8,10,14,18(22),19-heptaene-3-carboxamide | | Descriptor: | (3~{S},14~{E})-8-(azetidin-3-yl)-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, CHLORIDE ION, Cathepsin L1, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2017-11-17 | | Release date: | 2018-04-11 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
1WE5
 
 | | Crystal Structure of Alpha-Xylosidase from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative family 31 glucosidase yicI | | Authors: | Ose, T, Kitamura, M, Okuyama, M, Mori, H, Kimura, A, Watanabe, N, Yao, M, Tanaka, I. | | Deposit date: | 2004-05-24 | | Release date: | 2005-02-15 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Alpha-Xylosidase from Escherichia coli
TO BE PUBLISHED
|
|
1H8X
 
 | | Domain-swapped Dimer of a Human Pancreatic Ribonuclease Variant | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Canals, A, Pous, J, Guasch, A, Benito, A, Ribo, M, Vilanova, M, Coll, M. | | Deposit date: | 2001-02-16 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of an Engineered Domain-Swapped Ribonuclease Dimer and its Implications for the Evolution of Proteins Toward Oligomerization
Structure, 9, 2001
|
|
5N65
 
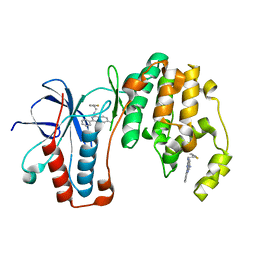 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9h | | Descriptor: | 2-phenyl-~{N}4-(2-thiophen-2-ylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5Y2T
 
 | | Structure of PPARgamma ligand binding domain - lobeglitazone complex | | Descriptor: | (5S)-5-[[4-[2-[[6-(4-methoxyphenoxy)pyrimidin-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Im, Y.J, Lee, M. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of PPAR gamma complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Sci Rep, 7, 2017
|
|
