3OND
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5C3Z
 
 | | Crystal structure of human ribokinase in complex with AMPPCP in C2 spacegroup | | Descriptor: | CHLORIDE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ribokinase, ... | | Authors: | Park, J, Chakrabarti, J, Singh, B, Gupta, R.S, Junop, M.S. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human ribokinase in complex with AMPPCP in C2 spacegroup
To Be Published
|
|
5C6F
 
 | | Crystal structures of ferritin mutants reveal side-on binding to diiron and end-on cleavage of oxygen | | Descriptor: | Bacterial non-heme ferritin, FE (III) ION, IMIDAZOLE | | Authors: | Kim, S, Kim, K.H, Seok, J.H, Park, Y.H, Jung, S.W, Chung, Y.B, Lee, D.B, Lee, J.H, Han, K.R, Cho, A.E, Lee, C, Chung, M.S. | | Deposit date: | 2015-06-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
5VM9
 
 | | Human Argonaute3 bound to guide RNA | | Descriptor: | Protein argonaute-3, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3'), RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3') | | Authors: | Park, M.S, Nakanishi, K. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Human Argonaute3 has slicer activity.
Nucleic Acids Res., 45, 2017
|
|
4R6B
 
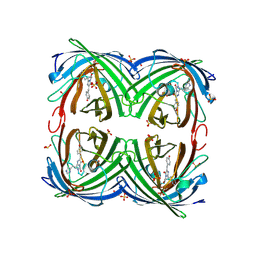 | | Rational Design of Enhanced Photoresistance in a Photoswitchable Fluorescent Protein | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Duan, C, Adam, V, Byrdin, M, Bourgeois, D. | | Deposit date: | 2014-08-23 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of enhanced photoresistance in a photoswitchable fluorescent protein.
Methods Appl Fluoresc, 3, 2015
|
|
7U2E
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADI-55688 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-55688 heavy chain, ADI-55688 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4R8T
 
 | | Structure of JEV protease | | Descriptor: | CHLORIDE ION, NS3, Serine protease subunit NS2B | | Authors: | Nair, D.T, Weinert, T, Wang, M, Olieric, V. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
3CMS
 
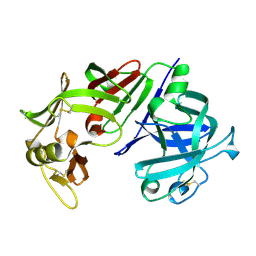 | | ENGINEERING ENZYME SUB-SITE SPECIFICITY: PREPARATION, KINETIC CHARACTERIZATION AND X-RAY ANALYSIS AT 2.0-ANGSTROMS RESOLUTION OF VAL111PHE SITE-MUTATED CALF CHYMOSIN | | Descriptor: | CHYMOSIN B | | Authors: | Newman, M, Frazao, C, Shearer, A, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1990-02-26 | | Release date: | 1992-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering enzyme subsite specificity: preparation, kinetic characterization, and X-ray analysis at 2.0-A resolution of Val111Phe site-mutated calf chymosin.
Biochemistry, 29, 1990
|
|
4RAK
 
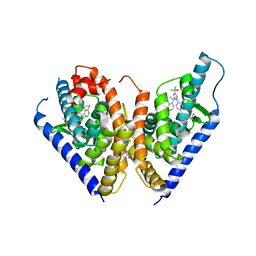 | |
4RFD
 
 | | Human carbonic anhydrase II in complex with 4-(4-sulfamoyl-phenoxy)-butylammonium | | Descriptor: | 4-(4-sulfamoyl-phenoxy)-butylammonium, BICARBONATE ION, Carbonic anhydrase 2, ... | | Authors: | Bozdag, M, Pinard, M.A, Carta, F, Masini, E, Scozzafava, A, Mckenna, R, Supuran, C.T. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | A class of 4-sulfamoylphenyl-omega-aminoalkyl ethers with effective carbonic anhydrase inhibitory action and antiglaucoma effects.
J.Med.Chem., 57, 2014
|
|
4R3M
 
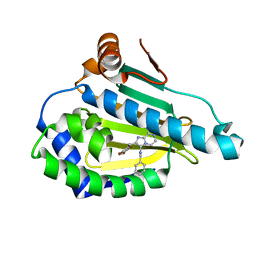 | | Crystal structure of Human Hsp90 with JR9 | | Descriptor: | Heat shock protein HSP 90-alpha, N~3~-benzyl-2-[(6-bromo-1,3-benzodioxol-5-yl)methyl]imidazo[1,2-a]pyrazine-3,8-diamine | | Authors: | Li, J, Yang, M, Ren, J, Xiong, B, He, J. | | Deposit date: | 2014-08-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multi-substituted 8-aminoimidazo[1,2-a]pyrazines by Groebke-Blackburn-Bienayme reaction and their Hsp90 inhibitory activity.
Org.Biomol.Chem., 13, 2015
|
|
5YYP
 
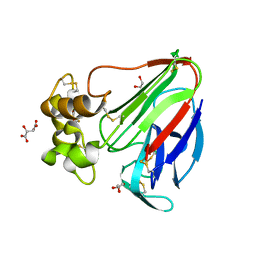 | | Structure K137A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5YYQ
 
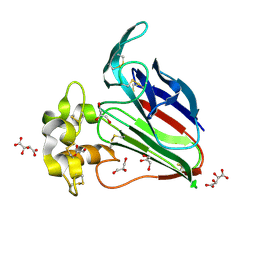 | | Structure K78A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
7P7O
 
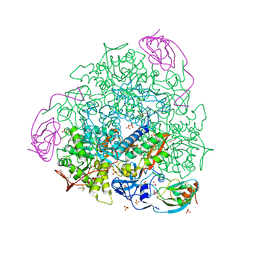 | | X-RAY CRYSTAL STRUCTURE OF SPOROSARCINA PASTEURII UREASE INHIBITED BY THE GOLD(I)-DIPHOSPHINE COMPOUND Au(PEt3)2Cl DETERMINED AT 1.87 ANGSTROMS | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Ciurli, S, Cianci, M, Messori, L, Massai, L. | | Deposit date: | 2021-07-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Medicinal Au(I) compounds targeting urease as prospective antimicrobial agents: unveiling the structural basis for enzyme inhibition.
Dalton Trans, 50, 2021
|
|
5YZA
 
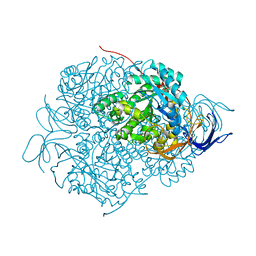 | | Crystal Structure of Human CRMP-2 with S522D mutation | | Descriptor: | Dihydropyrimidinase-related protein 2 | | Authors: | Sumi, T, Imasaki, T, Aoki, M, Sakai, N, Nitta, E, Shirouzu, M, Nitta, R. | | Deposit date: | 2017-12-13 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Altering Function of CRMP2 by Phosphorylation.
Cell Struct. Funct., 43, 2018
|
|
3CKS
 
 | | Urate oxidase complexed with 8-azaxanthine under 4.0 MPa oxygen pressure | | Descriptor: | 8-AZAXANTHINE, OXYGEN MOLECULE, SODIUM ION, ... | | Authors: | Colloc'h, N, Gabison, L, Chiadmi, M, Abraini, J.H, Prange, T. | | Deposit date: | 2008-03-17 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxygen pressurized X-ray crystallography: probing the dioxygen binding site in cofactorless urate oxidase and implications for its catalytic mechanism.
Biophys.J., 95, 2008
|
|
4RBX
 
 | |
4RFC
 
 | | Human carbonic anhydrase II in complex with tert-butyl 4-(4-sulfamoylphenoxy)butylcarbamate | | Descriptor: | Carbonic anhydrase 2, ZINC ION, tert-butyl 4-(4-sulfamoylphenoxy)butylcarbamate | | Authors: | Bozdag, M, Pinard, M.A, Carta, F, Masini, E, Scozzafava, A, Mckenna, R, Supuran, C.T. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | A class of 4-sulfamoylphenyl-omega-aminoalkyl ethers with effective carbonic anhydrase inhibitory action and antiglaucoma effects.
J.Med.Chem., 57, 2014
|
|
5YZ5
 
 | | Crystal Structure of Human CRMP-2 with T509D-T514D-S518D-S522D mutations | | Descriptor: | Dihydropyrimidinase-related protein 2, SULFATE ION | | Authors: | Imasaki, T, Sumi, T, Aoki, M, Sakai, N, Nitta, E, Shirouzu, M, Nitta, R. | | Deposit date: | 2017-12-13 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Altering Function of CRMP2 by Phosphorylation.
Cell Struct. Funct., 43, 2018
|
|
7OWB
 
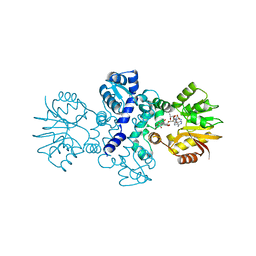 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with a region from 10-hydroxylase CalMB | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,5,7-trihydroxy-6,11-dioxo-4-{[2,3,6-trideoxy-3-(dimethylamino)-alpha-L-lyxo-hexopyranosyl]oxy}-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
4R1R
 
 | |
7OY1
 
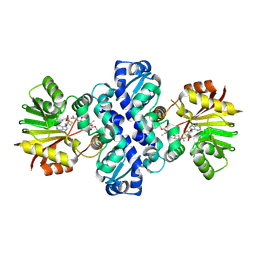 | | DnrK mutant RTCR | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,5,7-trihydroxy-6,11-dioxo-4-{[2,3,6-trideoxy-3-(dimethylamino)-alpha-L-lyxo-hexopyranosyl]oxy}-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7OVK
 
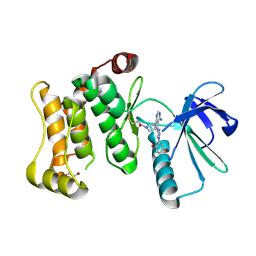 | | Protein kinase MKK7 in complex with 5-bromo-2-hydroxyphenyl-substituted pyrazolopyrimidine | | Descriptor: | 1-[(3~{R})-3-[4-azanyl-3-[1-(5-bromanyl-2-oxidanyl-phenyl)-1,2,3-triazol-4-yl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, Dual specificity mitogen-activated protein kinase kinase 7, GLYCEROL | | Authors: | Kleinboelting, S, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
7OVI
 
 | | Protein kinase MKK7 in complex with phenethyltriazole-substituted pyrazolopyrimidine | | Descriptor: | 1-[(3~{R})-3-[4-azanyl-3-[1-(2-phenylethyl)-1,2,3-triazol-4-yl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Kleinboelting, S, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
7OVM
 
 | | Protein kinase MKK7 in complex with cyclobutyl-substituted indazole | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[(1-cyclobutyl-1,2,3-triazol-4-yl)methyl]-3-(1~{H}-indazol-3-yl)-5-(propanoylamino)benzamide | | Authors: | Buehrmann, M, Wiese, J.N, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
