4ZNN
 
 | | MicroED structure of the segment, GVVHGVTTVA, from the A53T familial mutant of Parkinson's disease protein, alpha-synuclein residues 47-56 | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2015-05-05 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.41 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
4BUY
 
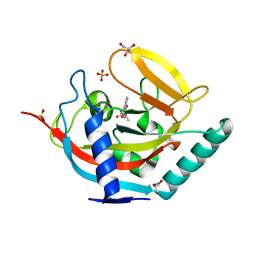 | | Crystal structure of human tankyrase 2 in complex with 5-methyl-5-(4-(4-oxo-3,4-dihydroquinazolin-2-yl)phenyl)imidazolidine-2,4-dione | | Descriptor: | (5S)-5-methyl-5-[4-(4-oxidanylidene-3H-quinazolin-2-yl)phenyl]imidazolidine-2,4-dione, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
2L1T
 
 | |
4BY2
 
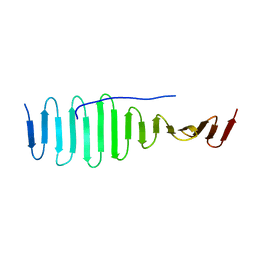 | | SAS-4 (dCPAP) TCP domain in complex with a Proline Rich Motif of Ana2 (dSTIL) of Drosophila Melanogaster | | Descriptor: | 1,2-ETHANEDIOL, ANASTRAL SPINDLE 2, SAS 4 | | Authors: | Cottee, M.A, Muschalik, N, Wong, Y.L, Johnson, C.M, Johnson, S, Andreeva, A, Oegema, K, Lea, S.M, Raff, J.W, van Breugel, M. | | Deposit date: | 2013-07-17 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly.
Elife, 2, 2013
|
|
8BR6
 
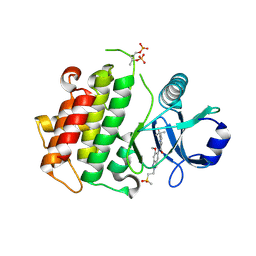 | | Discovery of IRAK4 Inhibitor 40 | | Descriptor: | ACETATE ION, Interleukin-1 receptor-associated kinase 4, ~{N}-[6-methoxy-2-(2-methylsulfonylethyl)-1,3-dihydroindazol-5-yl]-6-(2-oxidanylpropan-2-yl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wengner, A.M, Guimond, N, Thaler, T, Platzek, J, Ewerspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-11-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.167 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
4C98
 
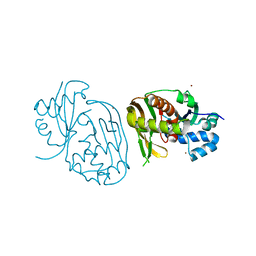 | |
2L13
 
 | |
8BR5
 
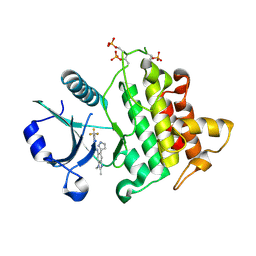 | | Discovery of IRAK4 Inhibitor 41 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-[2,3-dimethyl-6-(1~{H}-pyrazol-5-yl)benzimidazol-5-yl]-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-11-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
2L9D
 
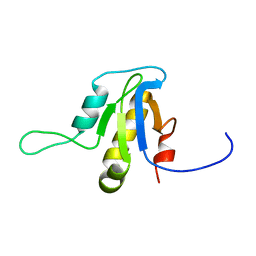 | | Solution structure of the protein YP_546394.1, the first structural representative of the pfam family PF12112 | | Descriptor: | Uncharacterized protein | | Authors: | Mohanty, B, Serrano, P, Geralt, M, Horst, R, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2011-02-08 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the protein YP_546394.1, the first structural representative of the pfam family PF12112
To be Published
|
|
8BR7
 
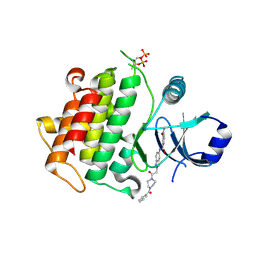 | | Discovery of IRAK4 Inhibitors BAY1834845 and BAY1830839 | | Descriptor: | 3-nitro-~{N}-[2-[2-oxidanylidene-2-[4-(phenylcarbonyl)piperazin-1-yl]ethyl]indazol-5-yl]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-11-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
4BSJ
 
 | | Crystal structure of VEGFR-3 extracellular domains D4-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 3 | | Authors: | Leppanen, V.-M, Tvorogov, D, Kisko, K, Prota, A.E, Jeltsch, M, Anisimov, A, Markovic-Mueller, S, Stuttfeld, E, Goldie, K.N, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Mechanistic Insights Into Vegfr-3 Ligand Binding and Activation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BSK
 
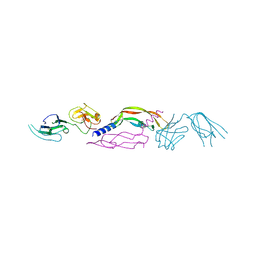 | | Crystal structure of VEGF-C in complex with VEGFR-3 domains D1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, VASCULAR ENDOTHELIAL GROWTH FACTOR C, ... | | Authors: | Leppanen, V.M, Tvorogov, D, Kisko, K, Prota, A.E, Jeltsch, M, Anisimov, A, Markovic-Mueller, S, Stuttfeld, E, Goldie, K.N, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.201 Å) | | Cite: | Structural and Mechanistic Insights Into Vegfr-3 Ligand Binding and Activation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BSU
 
 | | Structure of the ectodomain of LGR5 in complex with R-spondin-1 (Fu1Fu2) in C2 crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING G-PROTEIN COUPLED RECEPTOR 5, ... | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
2LK9
 
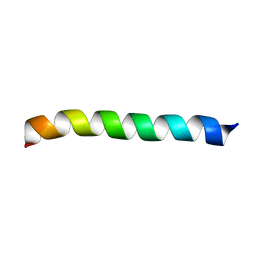 | | Structure of BST-2/Tetherin Transmembrane Domain | | Descriptor: | Bone marrow stromal antigen 2 | | Authors: | Skasko, M, Wang, Y, Tian, Y, Tokarev, A, Munguia, J, Ruiz, A, Stephens, E, Opella, S, Guatelli, J. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 Vpu Protein Antagonizes Innate Restriction Factor BST-2 via Lipid-embedded Helix-Helix Interactions.
J.Biol.Chem., 287, 2012
|
|
2LN8
 
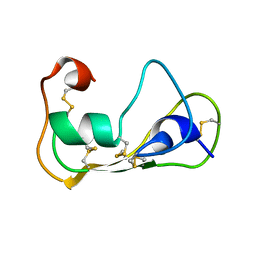 | | The solution structure of theromacin | | Descriptor: | Theromacin | | Authors: | Jung, S, Soennichsen, F.D, Hung, C.-W, Tholey, A, Boidin-Wichlacz, C, Hausgen, W, Gelhaus, C, Desel, C, Podschun, R, Watzig, V, Tasiemski, A, Leippe, M, Groetzinger, J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Macin family of antimicrobial proteins combines antimicrobial and nerve repair activities.
J.Biol.Chem., 287, 2012
|
|
4EG9
 
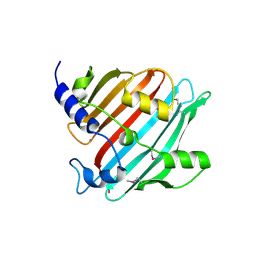 | | 1.9 Angstrom resolution crystal structure of Se-methionine hypothetical protein SAOUHSC_02783 from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Uncharacterized protein SAOUHSC_02783 | | Authors: | Biancucci, M, Minasov, G, Halavaty, A, Filippova, E.V, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-04-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom resolution crystal structure of Se-methionine hypothetical protein SAOUHSC_02783 from Staphylococcus aureus
TO BE PUBLISHED
|
|
4CDC
 
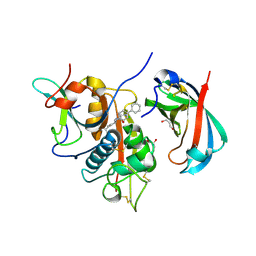 | | Human DPP1 in complex with (2S)-2-amino-N-((1S)-1-cyano-2-(4- phenylphenyl)ethyl)butanamide | | Descriptor: | (2S)-2-azanyl-N-[(2S)-1-azanylidene-3-(4-phenylphenyl)propan-2-yl]butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Debreczeni, J, Edman, K, Furber, M, Tiden, A, Gardiner, P, Mete, T, Ford, R, Millichip, I, Stein, L, Mather, A, Kinchin, E, Luckhurst, C, Cage, P, Sanghanee, H, Breed, J, Wissler, L. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cathepsin C Inhibitors: Property Optimization and Identification of a Clinical Candidate.
J.Med.Chem., 57, 2014
|
|
4CEM
 
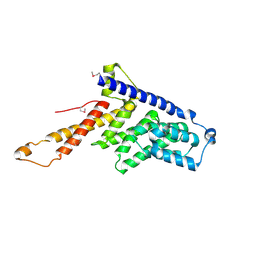 | | Crystal structure of the first MIF4G domain of human nonsense mediated decay factor UPF2 | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 2 | | Authors: | Clerici, M, Deniaud, A, Boehm, V, Gehring, N.H, Schaffitzel, C, Cusack, S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-12-11 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Analysis of the Three Mif4G Domains of Nonsense-Mediated Decay Factor Upf2.
Nucleic Acids Res., 42, 2014
|
|
4CD3
 
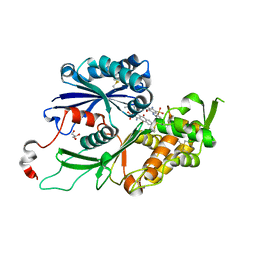 | | RnNTPDase2 X4 variant in complex with PSB-071 | | Descriptor: | 1-AMINO-4-(3-METHYLPHENYL)AMINO-9,10-DIOXO-9,10-DIHYDROANTHRACENE-2-SULFONATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, GLYCEROL | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2013-10-29 | | Release date: | 2014-02-12 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Ntpdase2 in Complex with the Sulfoanthraquinone Inhibitor Psb-071.
J.Struct.Biol., 185, 2014
|
|
4BOU
 
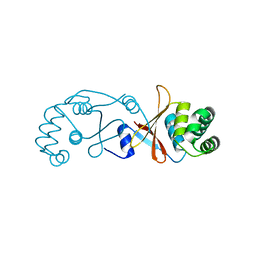 | | Structure of OTUD3 OTU domain | | Descriptor: | OTU DOMAIN-CONTAINING PROTEIN 3 | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
4BME
 
 | | Crystal structure of the N terminal domain of human Galectin 8, F19Y mutant | | Descriptor: | GALECTIN-8, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Buzamet, E, Ruiz, F.M, Menendez, M, Romero, A, Gabius, H.J, Solis, D. | | Deposit date: | 2013-05-07 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Natural Single Amino Acid Polymorphism (F19Y) in Human Galectin-8: Detection of Structural Alterations and Increased Growth-Regulatory Activity on Tumor Cells.
FEBS J., 281, 2014
|
|
2MQK
 
 | |
4BXZ
 
 | | RNA Polymerase II-Bye1 complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-16 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2MPM
 
 | | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: the N-terminal Region of CCR3 Bound to CCL11/Eotaxin-1 | | Descriptor: | CCR3, Eotaxin | | Authors: | Millard, C.J, Ludeman, J.P, Canals, M, Bridgford, J.L, Hinds, M.G, Clayton, D.J, Christopoulos, A, Payne, R.J, Stone, M.J. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-10 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: The N-Terminal Region of CCR3 Bound to CCL11/Eotaxin-1.
Structure, 22, 2014
|
|
2MRZ
 
 | | Dimeric structure of the Human A-box | | Descriptor: | DNA (5'-D(P*TP*CP*CP*TP*TP*TP*TP*CP*CP*A)-3') | | Authors: | Garavis, M, Escaja, N, Gabelica, V, Villasante, A, Gonzalez, C. | | Deposit date: | 2014-07-18 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Centromeric Alpha-Satellite DNA Adopts Dimeric i-Motif Structures Capped by AT Hoogsteen Base Pairs.
Chemistry, 21, 2015
|
|
