1NGK
 
 | | Crystallographic Structure of Mycobacterium tuberculosis Hemoglobin O | | Descriptor: | CYANIDE ION, Hemoglobin-like protein HbO, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Milani, M, Savard, P.-Y, Oullet, H, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2002-12-17 | | Release date: | 2003-05-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A TyrCD1/TrpG8 hydrogen bond network and a TyrB10-TyrCD1 covalent link shape the heme distal site of Mycobacterium tuberculosis hemoglobin O
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NHZ
 
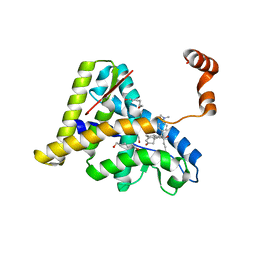 | | Crystal Structure of the Antagonist Form of Glucocorticoid Receptor | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, GLUCOCORTICOID RECEPTOR, HEXANE-1,6-DIOL | | Authors: | Kauppi, B, Jakob, C, Farnegardh, M, Yang, J, Ahola, H, Alarcon, M, Calles, K, Engstrom, O, Harlan, J, Muchmore, S, Ramqvist, A.-K, Thorell, S, Ohman, L, Greer, J, Gustafsson, J.-A, Carlstedt-Duke, J, Carlquist, M. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structures of Antagonistic and Agonistic Forms of the Glucocorticoid Receptor Ligand-binding Domain:
RU-486 INDUCES A TRANSCONFORMATION THAT LEADS TO ACTIVE ANTAGONISM.
J.Biol.Chem., 278, 2003
|
|
4Y7E
 
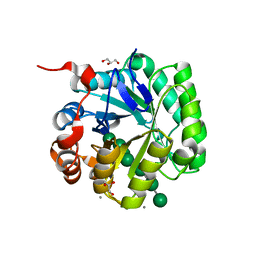 | | Crystal structure of beta-mannanase from Streptomyces thermolilacinus with mannohexaose | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kumagai, Y, Yamashita, K, Okuyama, M, Hatanaka, T, Yao, M, Kimura, A. | | Deposit date: | 2015-02-14 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition
Febs J., 282, 2015
|
|
7JKM
 
 | |
7JLZ
 
 | |
3FV1
 
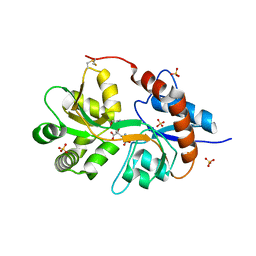 | | Crystal Structure of the human glutamate receptor, GluR5, ligand-binding core in complex with dysiherbaine in space group P1 | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
4DFL
 
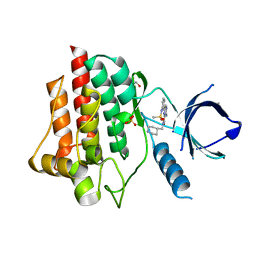 | | Crystal structure of spleen tyrosine kinase complexed with a sulfonamidopyrazine piperidine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-amino-6-{3-[(methylsulfonyl)amino]phenyl}-N-(piperidin-4-ylmethyl)pyrazine-2-carboxamide, SULFATE ION, ... | | Authors: | Lopez, M, Segarra, V, Vidal, B, Wenzkowski, C, Jestel, A, Krapp, S, Blaesse, M, Nagel, S, Schreiner, P. | | Deposit date: | 2012-01-24 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Pyrazine-based Syk kinase inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4BEG
 
 | |
4AYL
 
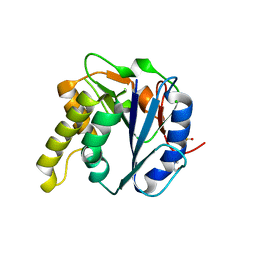 | | Molecular structure of a metal-independent bacterial glycosyltransferase that catalyzes the synthesis of histo-blood group A antigen | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BOGT-METAL-INDEPENDENT GLYCOSYLTRANSFERASE, CALCIUM ION, ... | | Authors: | Thiyagarajan, N, Pham, T.T.K, Stinsonb, B, Sundriyala, A, Tumbale, P, Lizotte-Waniewskib, M, Brewb, K, Acharya, K.R. | | Deposit date: | 2012-06-21 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structure of a Metal-Independent Bacterial Glycosyltransferase that Catalyzes the Synthesis of Histo-Blood Group a Antigen
Sci.Rep., 2, 2012
|
|
4YN3
 
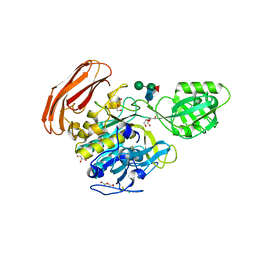 | | Crystal structure of Cucumisin complex with pro-peptide | | Descriptor: | CHLORIDE ION, Cucumisin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murayama, K, Kato-Murayama, M, Yokoyama, S, Arima, K, Shirouzu, M. | | Deposit date: | 2015-03-09 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of cucumisin protease activity regulation by its propeptide
J. Biochem., 161, 2017
|
|
8AQH
 
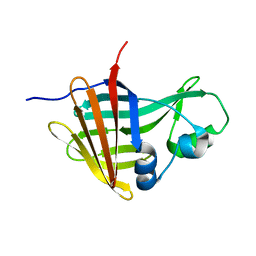 | | NanoLuc-Y94A luciferase mutant | | Descriptor: | NanoLuc luciferase | | Authors: | Nemergut, M, Marek, M. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Illuminating the mechanism and allosteric behavior of NanoLuc luciferase.
Nat Commun, 14, 2023
|
|
8AQ6
 
 | |
8AQI
 
 | |
7K0T
 
 | |
7K0S
 
 | |
4G9J
 
 | | Protein Ser/Thr phosphatase-1 in complex with cell-permeable peptide | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, synthetic peptide | | Authors: | Sukackaite, R, Chatterjee, J, Beullens, M, Bollen, M, Koehn, M, Hart, D.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of a Peptide that Selectively Activates Protein Phosphatase-1 in Living Cells.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH CHLORALHYDRATE | | Descriptor: | PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION, TRI-CHLORO-ACETALDEHYDE | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
4ABT
 
 | | Crystal structure of Type IIF restriction endonuclease NgoMIV with cognate uncleaved DNA | | Descriptor: | 5'-D(*TP*GP*CP*GP*CP*CP*GP*GP*CP*GP*CP)-3', CALCIUM ION, TYPE-2 RESTRICTION ENZYME NGOMIV | | Authors: | Manakova, E.N, Grazulis, S, Zaremba, M, Tamulaitiene, G, Golovenko, D, Siksnys, V. | | Deposit date: | 2011-12-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure of Type Iif Restriction Endonuclease Ngomiv with Cognate Uncleaved DNA
To be Published
|
|
5NUK
 
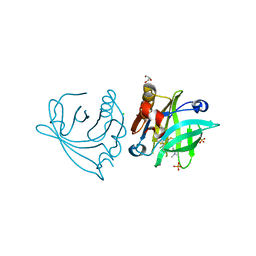 | | Engineered beta-lactoglobulin: variant I56F-L39A-M107F in complex with chlorpromazine (LG-FAF-CLP) | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Beta-lactoglobulin, CHLORIDE ION, ... | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Lazinska, I, Szydlowska, J, Lewinski, K. | | Deposit date: | 2017-04-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The engineered beta-lactoglobulin with complementarity to the chlorpromazine chiral conformers.
Int. J. Biol. Macromol., 114, 2018
|
|
7JRK
 
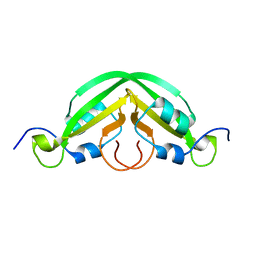 | |
5O1U
 
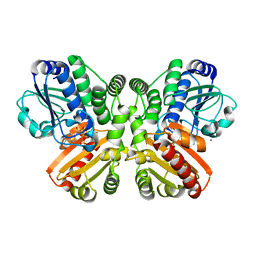 | | Structure of wildtype T.maritima PDE (TM1595) with AMP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-05-19 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
7K1U
 
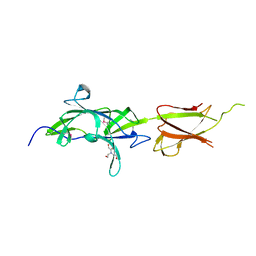 | | Crystal Structure of SrtB-anchored Collagen-binding Adhesin Fragment (residues 206-565) from Clostridioides difficile strain 630 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Collagen-binding Adhesin | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of SrtB-anchored Collagen-binding Adhesin Fragment (residues 206-565) from Clostridioides difficile strain 630
To Be Published
|
|
1LHK
 
 | | ROLE OF PROLINE RESIDUES IN HUMAN LYSOZYME STABILITY: A SCANNING CALORIMETRIC STUDY COMBINED WITH X-RAY STRUCTURE ANALYSIS OF PROLINE MUTANTS | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Herning, T, Kuroki, R, Yutani, K, Kikuchi, M. | | Deposit date: | 1992-03-27 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of proline residues in human lysozyme stability: a scanning calorimetric study combined with X-ray structure analysis of proline mutants.
Biochemistry, 31, 1992
|
|
4YST
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 24.9 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
7KCZ
 
 | | CRYSTAL STRUCTURE OF RHESUS MACAQUE (MACACA MULATTA) IGG1 FC FRAGMENT- FC-GAMMA RECEPTOR III COMPLEX V158 MUTANT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG1 Fc, ... | | Authors: | Tolbert, W.D, Pazgier, M. | | Deposit date: | 2020-10-07 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Decoding human-macaque interspecies differences in Fc-effector functions: The structural basis for CD16-dependent effector function in Rhesus macaques.
Front Immunol, 13, 2022
|
|
