7CDK
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 42 kGy (4500 images from 1st half of data set) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDO
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 21 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
1N3T
 
 | | Biosynthesis of pteridins. Reaction mechanism of GTP cyclohydrolase I | | Descriptor: | GTP cyclohydrolase I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Rebelo, J, Auerbach, G, Bader, G, Bracher, A, Nar, H, Hoesl, C, Schramek, N, Kaiser, J, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biosynthesis of Pteridines. Reaction Mechanism of GTP Cyclohydrolase I
J.MOL.BIOL., 326, 2003
|
|
7CDU
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 1700 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDT
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 830 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
1MTZ
 
 | | Crystal Structure of the Tricorn Interacting Factor F1 | | Descriptor: | Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
3VBZ
 
 | | Crystal structure of Taipoxin beta subunit isoform 2 | | Descriptor: | Phospholipase A2 homolog, taipoxin beta chain | | Authors: | Cendron, L, Micetic, I, Polverino, P, Beltramini, M, Paoli, M. | | Deposit date: | 2012-01-03 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis of trimeric phospholipase A(2) neurotoxin from the Australian taipan snake venom.
Febs J., 279, 2012
|
|
5U3W
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 7 | | Descriptor: | 6-(2-{[([1,1'-biphenyl]-4-carbonyl)(cyclopropyl)amino]methyl}phenoxy)hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1MWD
 
 | | WILD TYPE DEOXY MYOGLOBIN | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, A.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-11 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
1MWO
 
 | | Crystal Structure Analysis of the Hyperthermostable Pyrocoocus woesei alpha-amylase | | Descriptor: | CALCIUM ION, ZINC ION, alpha amylase | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-09-30 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
5UMG
 
 | | Crystal structure of dihydropteroate synthase from Klebsiella pneumoniae subsp. | | Descriptor: | Dihydropteroate synthase, GLYCEROL | | Authors: | Chang, C, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-27 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of dihydropteroate synthase from Klebsiella pneumoniae subsp.
To Be Published
|
|
1MY9
 
 | | Solution structure of a K+ cation stabilized dimeric RNA quadruplex containing two G:G(:A):G:G(:A) hexads, G:G:G:G tetrads and UUUU loops | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*UP*UP*UP*UP*GP*GP*AP*GP*G)-3' | | Authors: | Liu, H, Matsugami, A, Katahira, M, Uesugi, S. | | Deposit date: | 2002-10-04 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Dimeric RNA Quadruplex Architecture Comprised of Two G:G(:A):G:G(:A) Hexads, G:G:G:G Tetrads and UUUU Loops
J.Mol.Biol., 322, 2002
|
|
5V2O
 
 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | Descriptor: | 1,3,5-BENZENETRICARBOXYLIC ACID, GLYCEROL, NONAETHYLENE GLYCOL, ... | | Authors: | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | Deposit date: | 2017-03-06 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7CB0
 
 | |
1TET
 
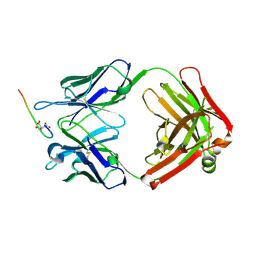 | |
7CB5
 
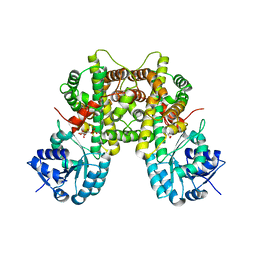 | |
5V47
 
 | | Crystal structure of the SR1 domain of lizard sacsin | | Descriptor: | Lizard sacsin, SULFATE ION | | Authors: | Pan, T, Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
7CB6
 
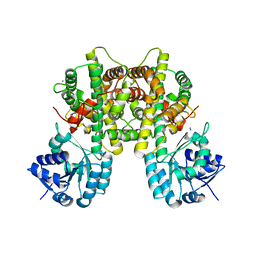 | |
5UQS
 
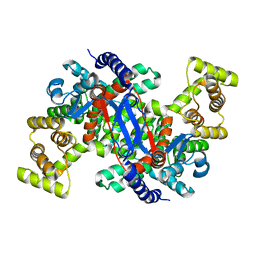 | |
1N3E
 
 | | Crystal structure of I-CreI bound to a palindromic DNA sequence I (palindrome of left side of wildtype DNA target sequence) | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*C)-3', 5'-D(P*GP*AP*CP*GP*TP*TP*TP*TP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Turmel, M, Lemieux, C, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2002-10-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexible DNA Target Site Recognition by Divergent Homing Endonuclease Isoschizomers I-CreI and I-MsoI
J.Mol.Biol., 329, 2003
|
|
1TQ1
 
 | | Solution structure of At5g66040, a putative protein from Arabidosis Thaliana | | Descriptor: | senescence-associated family protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Singh, S, Lee, M.S, Tyler, E.M, Shahan, M.N, Vinarov, D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-16 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a single-domain thiosulfate sulfurtransferase from Arabidopsis thaliana.
Protein Sci., 15, 2006
|
|
1N0X
 
 | | Crystal Structure of a Broadly Neutralizing Anti-HIV-1 Antibody in Complex with a Peptide Mimotope | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, B2.1 peptide, GLYCEROL, ... | | Authors: | Saphire, E.O, Montero, M, Menendez, A, Irving, M.B, Zwick, M.B, Parren, P.W.H.I, Burton, D.R, Scott, J.K, Wilson, I.A. | | Deposit date: | 2002-10-15 | | Release date: | 2004-04-13 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Broadly Neutralizing Anti-HIV-1 Antibody in Complex with a Peptide Mimotope
To be Published
|
|
5V75
 
 | |
5V7V
 
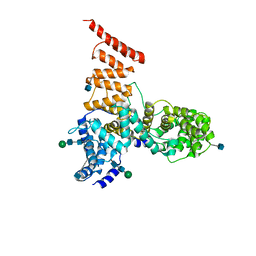 | | Cryo-EM structure of ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ERAD-associated E3 ubiquitin-protein ligase component HRD3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mi, W, Schoebel, S, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
1KFM
 
 | |
