8OYY
 
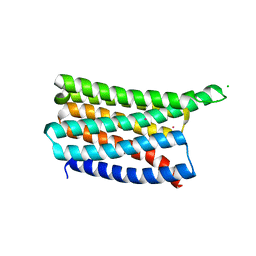 | | De novo designed soluble GPCR-like fold GLF_32 | | Descriptor: | CHLORIDE ION, De novo designed soluble GPCR-like protein, POTASSIUM ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
1BDA
 
 | |
4KHO
 
 | |
5M6I
 
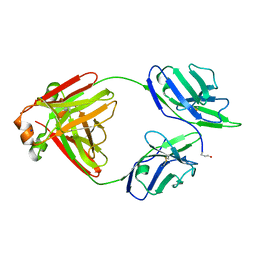 | | Crystal structure of non-cardiotoxic Bence-Jones light chain dimer M8 | | Descriptor: | SODIUM ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5M6Z
 
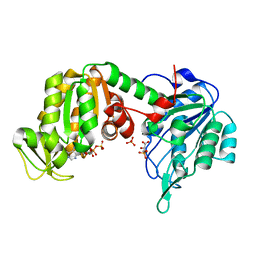 | | The X-ray structure of human M189I PGK-1 mutant in partially closed conformation | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ilari, A, Fiorillo, A, Petrosino, M, Cipollone, A. | | Deposit date: | 2016-10-26 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
5GG6
 
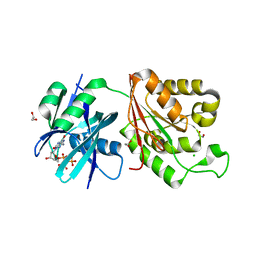 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP | | Descriptor: | 1,2-ETHANEDIOL, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GGB
 
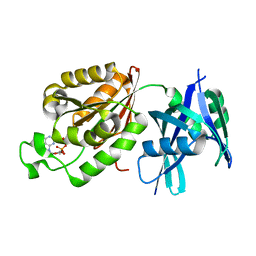 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGDP | | Descriptor: | 2'-deoxy-8-oxoguanosine 5'-(trihydrogen diphosphate), Hydrolase, NUDIX family protein | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2KMJ
 
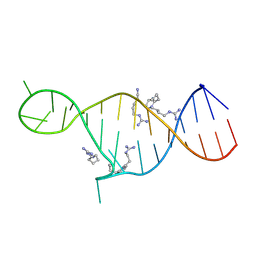 | | High resolution NMR solution structure of a complex of HIV-2 TAR RNA and a synthetic tripeptide in a 1:2 stoichiometry | | Descriptor: | Pyrimidinylpeptide, RNA (28-MER) | | Authors: | Ferner, J, Suhartono, M, Breitung, S, Jonker, H.R.A, Hennig, M, Woehnert, J, Goebel, M, Schwalbe, H. | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries.
Chembiochem, 10, 2009
|
|
7JP1
 
 | |
4C4G
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
1FLN
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: D58P REDUCED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-18 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
8OYW
 
 | |
3R6G
 
 | |
6WHF
 
 | | class C beta-lactamase from Escherichia coli in complex with cephalothin | | Descriptor: | (2R)-5-[(acetyloxy)methyl]-2-{(1R)-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | class C beta-lactamase from Escherichia coli in complex with Cephalothin
To Be Published
|
|
4KP0
 
 | | Crystal Structure of the human Chymase with TJK002 | | Descriptor: | 4-({1-[(4-methyl-1-benzothiophen-3-yl)methyl]-1H-benzimidazol-2-yl}sulfanyl)butanoic acid, Chymase | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2013-05-12 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a complex of human chymase with its benzimidazole derived inhibitor
J.SYNCHROTRON RADIAT., 20, 2013
|
|
1FLD
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57T OXIDIZED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-17 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
4KP7
 
 | | Structure of Plasmodium IspC in complex with a beta-thia-isostere derivative of Fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, MANGANESE (III) ION, ... | | Authors: | Kunfermann, A, Bacher, A, Groll, M. | | Deposit date: | 2013-05-13 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | IspC as Target for Antiinfective Drug Discovery: Synthesis, Enantiomeric Separation, and Structural Biology of Fosmidomycin Thia Isosters.
J.Med.Chem., 56, 2013
|
|
3R7S
 
 | | Crystal Structure of Apo Caspase2 | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18 | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
6WIF
 
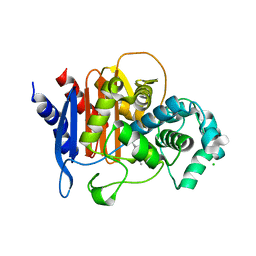 | | Class C beta-lactamase from Acinetobacter baumannii in complex with 4-(Ethyl(methyl)carbamoyl)phenyl boronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | class C beta-lactamase from Acinetobacter baumannii in complex with 4-(Ethyl(methyl)carbamoyl)phenyl boronic acid
To Be Published
|
|
2KNV
 
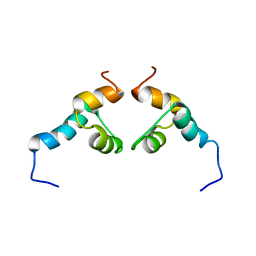 | |
6WIP
 
 | | Class A beta-lactamase from Micromonospora aurantiaca ATCC 27029 | | Descriptor: | ACETATE ION, Beta-lactamase, SULFATE ION | | Authors: | Chang, C, Tesar, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | class A beta-lactamase from
Micromonospora aurantiaca ATCC 27029
To Be Published
|
|
6YTD
 
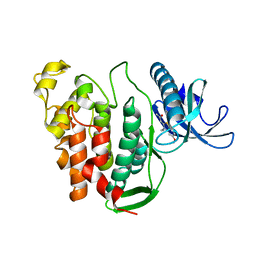 | | CLK1 V324A mutant bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6WJM
 
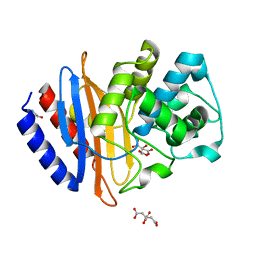 | | The crystal structure beta-lactamase from Desulfarculus baarsii DSM 2075 | | Descriptor: | ALANINE, Beta-lactamase, CITRIC ACID | | Authors: | Chang, C, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The crystal structure beta-lactamase from Desulfarculus baarsii DSM 2075
To Be Published
|
|
6WJT
 
 | | 2.0 Angstrom Resolution Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 in Complex with S-Adenosyl-L-Homocysteine | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6YTI
 
 | | CLK1 bound with ETH1610 (Cpd 17) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK1, methyl 9-[(2-fluoranyl-4-methoxy-phenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
