8B7S
 
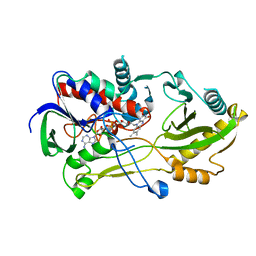 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
6OZ4
 
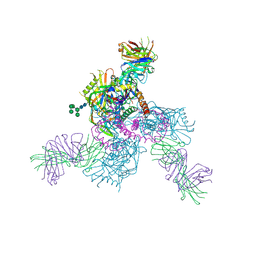 | |
8B8S
 
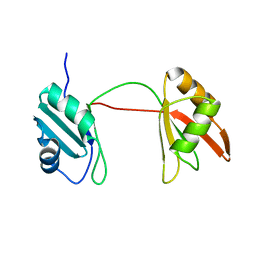 | | Solution structure of tandem RRM1 and RRM2 domains of yeast NPL3 | | Descriptor: | Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 | | Authors: | Kachariya, N, Sattler, M, Keil, P, Strasser, K. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res., 51, 2023
|
|
6RRA
 
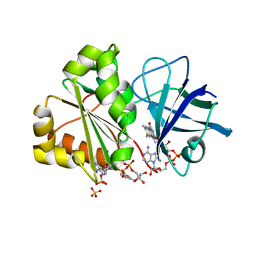 | | X-RAY STRUCTURE OF THE FERREDOXIN-NADP REDUCTASE FROM BRUCELLA OVIS IN COMPLEX WITH NADP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Martinez-Julvez, M, Taleb, V, Medina, M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Towards the competent conformation for catalysis in the ferredoxin-NADP+reductase from the Brucella ovis pathogen.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
6L9T
 
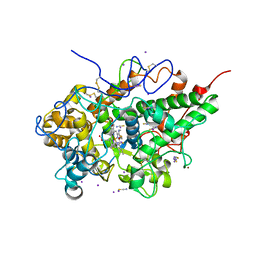 | | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Viswanathan, V, Pandey, N, Singh, A, Sinha, M, Singh, R.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution
To Be Published
|
|
4I4V
 
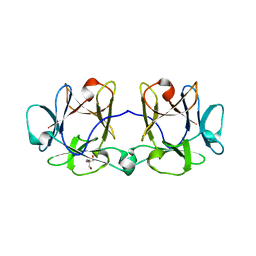 | | BEL beta-trefoil complex with N-acetylgalactosamine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, BEL beta-trefoil | | Authors: | Bovi, M, Cenci, L, Perduca, M, Capaldi, S, Carrizo, M.E, Civiero, L, Chiarelli, L.R, Galliano, M, Monaco, H.L. | | Deposit date: | 2012-11-28 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | BEL {beta}-trefoil: A novel lectin with antineoplastic properties in king bolete (Boletus edulis) mushrooms.
Glycobiology, 23, 2013
|
|
5UCI
 
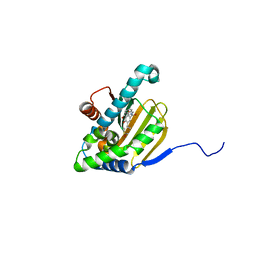 | | Hsp90b N-terminal domain with inhibitors | | Descriptor: | (2,4-Dihydroxy-3-(hydroxymethyl)-5-isopropylphenyl)(isoindolin-2-yl)methanone, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Peng, S, Balch, M, Matts, R, Deng, J. | | Deposit date: | 2016-12-22 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided design of an Hsp90 beta N-terminal isoform-selective inhibitor.
Nat Commun, 9, 2018
|
|
6F2G
 
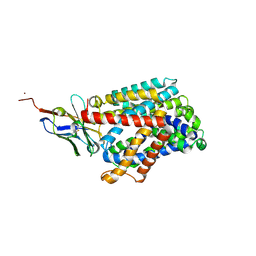 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | Nanobody 74, Putative amino acid/polyamine transport protein, ZINC ION | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
6RZU
 
 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
5JR5
 
 | | RHCC in Complex with Elemental Sulfur | | Descriptor: | SULFATE ION, Tetrabrachion, octathiocane | | Authors: | McDougall, M, Meier, M, Stetefeld, J. | | Deposit date: | 2016-05-05 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Archaea S-layer nanotube from a "black smoker" in complex with cyclo-octasulfur (S8 ) rings.
Proteins, 85, 2017
|
|
7DED
 
 | | Mevo lectin complex with mannoheptose (Man7) | | Descriptor: | 1,2-ETHANEDIOL, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, ... | | Authors: | Sivaji, N, Surolia, A, Vijayan, M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.228 Å) | | Cite: | Mevo lectin specificity toward high-mannose structures with terminal alpha Man(1,2) alpha Man residues and its implication to inhibition of the entry of Mycobacterium tuberculosis into macrophages.
Glycobiology, 31, 2021
|
|
6AWT
 
 | | Structure of PR 10 Allergen in complex with epicatechin | | Descriptor: | (2R,3R)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol, BENZOIC ACID, PR 10 protein, ... | | Authors: | Offermann, L.R, Perdue, M, McBride, J, Hurlburt, B.K, Maleki, S.J, Pote, S.S, Chruszcz, M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of PR-10 Allergen Ara h 8.01.
To Be Published
|
|
8BD5
 
 | | Cas12k-sgRNA-dsDNA-S15-TniQ-TnsC transposon recruitment complex | | Descriptor: | 30S ribosomal protein S15, ADENOSINE-5'-TRIPHOSPHATE, DNA non-target strand, ... | | Authors: | Schmitz, M, Querques, I, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
8RZ6
 
 | | SeMet derivative structure of the condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase | | Descriptor: | FORMIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Karanth, M, Schmelz, S, Kirkpatrick, J, Krausze, J, Scrima, A, Carlomagno, T. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain.
Sci Adv, 10, 2024
|
|
8GEW
 
 | | H-FABP crystal soaked in a bromo palmitic acid solution | | Descriptor: | 2-Bromopalmitic acid, Fatty acid-binding protein, heart, ... | | Authors: | Howard, E, Cousido-Siah, A, Alvarez, A, Espinosa, Y, Podjarny, A, Mitschler, A, Carlevaro, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-30 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Lipid exchange in crystal-confined fatty acid binding proteins: X-ray evidence and molecular dynamics explanation.
Proteins, 91, 2023
|
|
6F2W
 
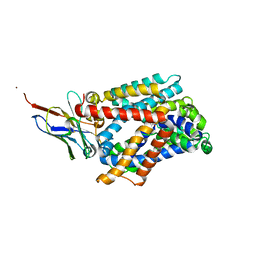 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | ALPHA-AMINOISOBUTYRIC ACID, Nanobody 74, Putative amino acid/polyamine transport protein, ... | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
8BD4
 
 | | TniQ-capped Tns-ATP-dsDNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*C)-3'), MAGNESIUM ION, ... | | Authors: | Querques, I, Schmitz, M, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
4IKP
 
 | | Crystal structure of coactivator-associated arginine methyltransferase 1 with methylenesinefungin | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Dombrovski, L, He, H, Ibanez, G, Wernimont, A, Zheng, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-27 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
8JV7
 
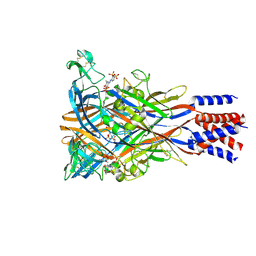 | | Cryo-EM structure of the panda P2X7 receptor in complex with PPADS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(E)-[4-methanoyl-6-methyl-5-oxidanyl-3-(phosphonooxymethyl)pyridin-2-yl]diazenyl]benzene-1,3-disulfonic acid, P2X purinoceptor | | Authors: | Sheng, D, Hattori, M. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the orthosteric inhibition of P2X receptors by non-ATP analog antagonists.
Elife, 12, 2024
|
|
6ETZ
 
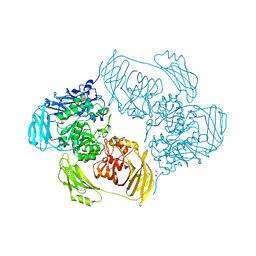 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Situ Random Microseeding and Streak Seeding Used for Growth of Crystals of Cold-Adapted Beta-D-Galactosidases: Crystal Structure of BetaDG from Arthrobacter sp. 32cB
Crystals, 8, 2018
|
|
5UCH
 
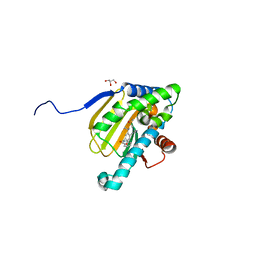 | | Hsp90b N-terminal domain with inhibitors | | Descriptor: | 2-(5-Hydroxy-4-(isoindoline-2-carbonyl)-2-isopropylphenyl)acetonitrile, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Peng, S, Balch, M, Matts, R, Deng, J. | | Deposit date: | 2016-12-22 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Structure-guided design of an Hsp90 beta N-terminal isoform-selective inhibitor.
Nat Commun, 9, 2018
|
|
7D56
 
 | |
7D4Y
 
 | |
8JV8
 
 | | Cryo-EM structure of the panda P2X7 receptor in complex with PPNDS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, P2X purinoceptor | | Authors: | Sheng, D, Hattori, M. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural insights into the orthosteric inhibition of P2X receptors by non-ATP analog antagonists.
Elife, 12, 2024
|
|
