6MWV
 
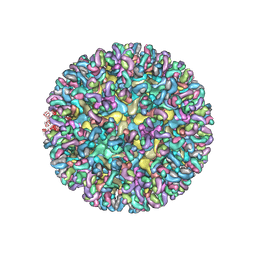 | | CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-58 Antibody | | Descriptor: | E1, E2, EEEV-58 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
7O0T
 
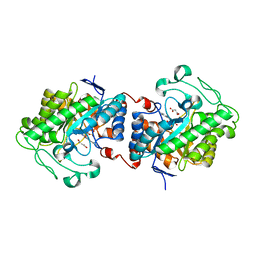 | | Crystal structure of Chloroflexus aggregans ene-reductase CaOYE holoenzyme | | Descriptor: | FLAVIN MONONUCLEOTIDE, MALONATE ION, NADH:flavin oxidoreductase/NADH oxidase | | Authors: | Robescu, M.S, Niero, M, Loprete, G, Bergantino, E, Cendron, L. | | Deposit date: | 2021-03-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A New Thermophilic Ene-Reductase from the Filamentous Anoxygenic Phototrophic Bacterium Chloroflexus aggregans .
Microorganisms, 9, 2021
|
|
6MX4
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6NEO
 
 | |
6MWC
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-5 antibody | | Descriptor: | E1, E2, EEEV-5 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6NZN
 
 | | Dimer-of-dimer amyloid fibril structure of glucagon | | Descriptor: | Glucagon | | Authors: | Gelenter, M.D, Smith, K.J, Liao, S.Y, Mandala, V.S, Dregni, A.J, Lamm, M.S, Tian, Y, Wei, X, Pochan, D.J, Tucker, T.J, Su, Y, Hong, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The peptide hormone glucagon forms amyloid fibrils with two coexisting beta-strand conformations.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1SLH
 
 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and dUDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1U1Z
 
 | | The Structure of (3R)-hydroxyacyl-ACP dehydratase (FabZ) | | Descriptor: | (3R)-hydroxymyristoyl-[acyl carrier protein] dehydratase, SULFATE ION | | Authors: | Kimber, M.S, Martin, F, Lu, Y, Houston, S, Vedadi, M, Dharamsi, A, Fiebig, K.M, Schmid, M, Rock, C.O. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa
J.Biol.Chem., 279, 2004
|
|
6JKI
 
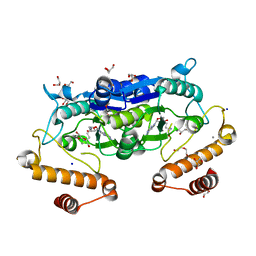 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 25, 2019
|
|
2ANQ
 
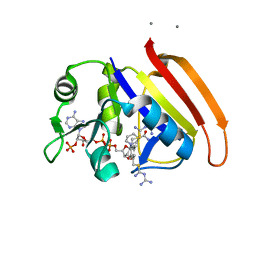 | | Crystal Structure of E.coli DHFR in complex with NADPH and the inhibitor compound 10a. | | Descriptor: | (2,5-dimethylbenzene-1,4-diyl)dimethanediyl bis(N-carbamimidoylcarbamimidothioate), Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
1KMG
 
 | | The Solution Structure Of Monomeric Copper-free Superoxide Dismutase | | Descriptor: | Superoxide Dismutase, ZINC ION | | Authors: | Banci, L, Bertini, I, Cantini, F, D'Onofrio, M, Viezzoli, M.S. | | Deposit date: | 2001-12-15 | | Release date: | 2002-10-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of copper-free SOD: The protein before binding copper.
Protein Sci., 11, 2002
|
|
4AIO
 
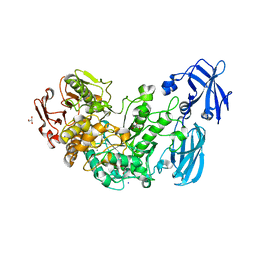 | | Crystal structure of the starch debranching enzyme barley limit dextrinase | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Moeller, M.S, Abou Hachem, M, Svensson, B, Henriksen, A. | | Deposit date: | 2012-02-11 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Starch-Debranching Enzyme Barley Limit Dextrinase Reveals Homology of the N-Terminal Domain to Cbm21.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2ANO
 
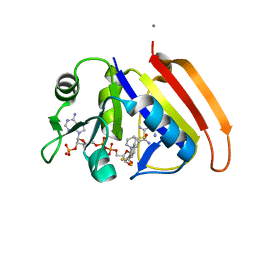 | | Crystal structure of E.coli dihydrofolate reductase in complex with NADPH and the inhibitor MS-SH08-17 | | Descriptor: | 1-{[N-(1-IMINO-GUANIDINO-METHYL)]SULFANYLMETHYL}-3-TRIFLUOROMETHYL-BENZENE, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
4V5T
 
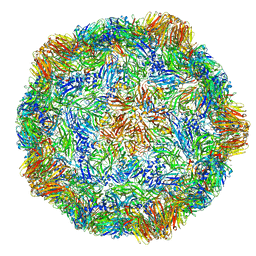 | | X-ray structure of the Grapevine Fanleaf virus | | Descriptor: | COAT PROTEIN | | Authors: | Schellenberger, P, Sauter, C, Lorber, B, Bron, P, Trapani, S, Bergdoll, M, Marmonier, A, Schmitt-Keichinger, C, Lemaire, O, Demangeat, G, Ritzenthaler, C. | | Deposit date: | 2011-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights Into Viral Determinants of Nematode Mediated Grapevine Fanleaf Virus Transmission.
Plos Pathog., 7, 2011
|
|
6I68
 
 | |
8DZF
 
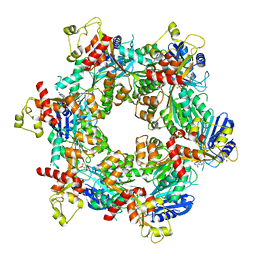 | | Cryo-EM structure of bundle-forming pilus extension ATPase from E.coli in the presence of AMP-PNP (class-2) | | Descriptor: | BfpD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ZINC ION | | Authors: | Nayak, A.R, Zhao, J, Donnenberg, M.S, Samso, M. | | Deposit date: | 2022-08-07 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Cryo-EM Structure of the Type IV Pilus Extension ATPase from Enteropathogenic Escherichia coli.
Mbio, 13, 2022
|
|
8DZE
 
 | | Cryo-EM structure of bundle-forming pilus extension ATPase from E. coli in the presence of AMP-PNP (class-1) | | Descriptor: | BfpD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ZINC ION | | Authors: | Nayak, A.R, Zhao, J, Donnenberg, M.S, Samso, M. | | Deposit date: | 2022-08-07 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM Structure of the Type IV Pilus Extension ATPase from Enteropathogenic Escherichia coli.
Mbio, 13, 2022
|
|
8DZG
 
 | | Cryo-EM structure of bundle-forming pilus extension ATPase from E.coli in the presence of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BfpD, MAGNESIUM ION, ... | | Authors: | Nayak, A.R, Zhao, J, Donnenberg, M.S, Samso, M. | | Deposit date: | 2022-08-07 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structure of the Type IV Pilus Extension ATPase from Enteropathogenic Escherichia coli.
Mbio, 13, 2022
|
|
8C9X
 
 | | human alpha7 nicotinic receptor in complex with the C4 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody C4, ... | | Authors: | Prevost, M.S, Barilone, N, Dejean de la Batie, G, Pons, S, Ayme, G, England, P, Gielen, M, Bontems, F, Pehau-Arnaudet, G, Maskos, U, Lafaye, P, Corringer, P.-J. | | Deposit date: | 2023-01-23 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | An original potentiating mechanism revealed by the cryo-EM structures of the human alpha 7 nicotinic receptor in complex with nanobodies.
Nat Commun, 14, 2023
|
|
8CAU
 
 | | human alpha7 nicotinic receptor in complex with the C4 nanobody and nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Prevost, M.S, Barilone, N, Dejean de la Batie, G, Pons, S, Ayme, G, England, P, Gielen, M, Bontems, F, Pehau-Arnaudet, G, Maskos, U, Lafaye, P, Corringer, P.-J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An original potentiating mechanism revealed by the cryo-EM structures of the human alpha 7 nicotinic receptor in complex with nanobodies.
Nat Commun, 14, 2023
|
|
8CI2
 
 | | human alpha7 nicotinic receptor in complex with the C4 nanobody under sub-saturating conditions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody C4, ... | | Authors: | Prevost, M.S, Barilone, N, Dejean de la Batie, G, Pons, S, Ayme, G, England, P, Gielen, M, Bontems, F, Pehau-Arnaudet, G, Maskos, U, Lafaye, P, Corringer, P.-J. | | Deposit date: | 2023-02-08 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | An original potentiating mechanism revealed by the cryo-EM structures of the human alpha 7 nicotinic receptor in complex with nanobodies.
Nat Commun, 14, 2023
|
|
8CE4
 
 | | Human alpha7 nicotinic receptor in complex with the E3 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody E3, ... | | Authors: | Prevost, M.S, Barilone, N, Dejean de la Batie, G, Pons, S, Ayme, G, England, P, Gielen, M, Bontems, F, Pehau-Arnaudet, G, Maskos, U, Lafaye, P, Corringer, P.-J. | | Deposit date: | 2023-02-01 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An original potentiating mechanism revealed by the cryo-EM structures of the human alpha 7 nicotinic receptor in complex with nanobodies.
Nat Commun, 14, 2023
|
|
8CI1
 
 | | Human alpha7 nicotinic receptor in complex with the E3 nanobody and nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Prevost, M.S, Barilone, N, Dejean de la Batie, G, Pons, S, Ayme, G, England, P, Gielen, M, Bontems, F, Pehau-Arnaudet, G, Maskos, U, Lafaye, P, Corringer, P.-J. | | Deposit date: | 2023-02-08 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | An original potentiating mechanism revealed by the cryo-EM structures of the human alpha 7 nicotinic receptor in complex with nanobodies.
Nat Commun, 14, 2023
|
|
6I41
 
 | |
6I5P
 
 | |
