6QH2
 
 | |
3TRQ
 
 | | Crystal structure of native rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Glycosylation of Skeletal Calsequestrin: IMPLICATIONS FOR ITS FUNCTION.
J.Biol.Chem., 287, 2012
|
|
3TZB
 
 | | Quinone Oxidoreductase (NQ02) bound to NSC13000 | | Descriptor: | 9-AMINOACRIDINE, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Dunstan, M.S, Leys, D. | | Deposit date: | 2011-09-27 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1901 Å) | | Cite: | In silico screening reveals structurally diverse, nanomolar inhibitors of NQO2 that are functionally active in cells and can modulate NF-kappa B signaling.
Mol.Cancer Ther., 11, 2012
|
|
6QFP
 
 | |
6WBX
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 1 | | Descriptor: | CALCIUM ION, DUF3367 domain-containing protein | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6WBY
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 2 | | Descriptor: | CALCIUM ION, DUF3367 domain-containing protein | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
1LZN
 
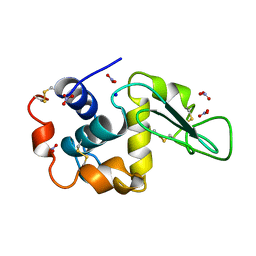 | | NEUTRON STRUCTURE OF HEN EGG-WHITE LYSOZYME | | Descriptor: | NITRATE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Bon, C.I, Lehmann, M.S, Wilkinson, C. | | Deposit date: | 1999-03-23 | | Release date: | 1999-04-01 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å) | | Cite: | Quasi-Laue neutron-diffraction study of the water arrangement in crystals of triclinic hen egg-white lysozyme.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1W0D
 
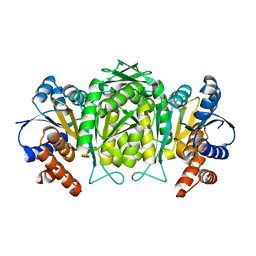 | | The high resolution structure of Mycobacterium tuberculosis LeuB (Rv2995c) | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | Authors: | Singh, R.K, Kefala, G, Janowski, R, Mueller-Dieckmann, C, Weiss, M.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-06-03 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The High Resolution Structure of Leub (Rv2995C) from Mycobacterium Tuberculosis
J.Mol.Biol., 346, 2005
|
|
3MMK
 
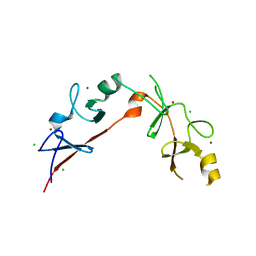 | | The structural basis for partial redundancy in a class of transcription factors, the lim-homeodomain proteins, in neural cell type specification | | Descriptor: | CHLORIDE ION, Fusion of LIM/homeobox protein Lhx4, linker, ... | | Authors: | Gadd, M.S, Langley, D.B, Guss, J.M, Matthews, J.M. | | Deposit date: | 2010-04-20 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | The structural basis for partial redundancy in a class of transcription factors, the lim-homeodomain proteins, in neural cell type specification.
J.Biol.Chem., 2011
|
|
3M13
 
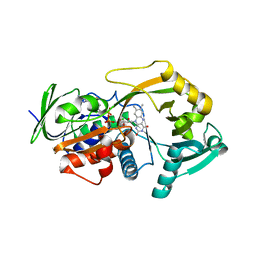 | | Crystal Structure of the Lys265Arg PEG-crystallized mutant of monomeric sarcosine oxidase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase, ... | | Authors: | Mathews, F.S, Chen, Z.-W, Jorns, M.S. | | Deposit date: | 2010-03-04 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of mutations at the oxygen activation site in monomeric sarcosine oxidase.
Biochemistry, 49, 2010
|
|
3TNU
 
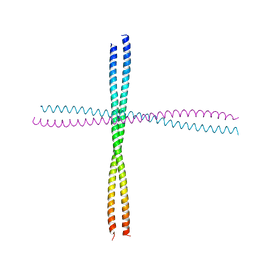 | | Heterocomplex of coil 2B domains of human intermediate filament proteins, keratin 5 (KRT5) and keratin 14 (KRT14) | | Descriptor: | Keratin, type I cytoskeletal 14, type II cytoskeletal 5 | | Authors: | Lee, C.H, Kim, M.S, Leahy, D.J, Coulombe, P.A. | | Deposit date: | 2011-09-02 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural basis for heteromeric assembly and perinuclear organization of keratin filaments.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5ITD
 
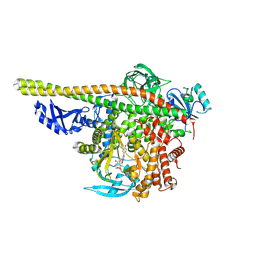 | | Crystal structure of PI3K alpha with PI3K delta inhibitor | | Descriptor: | 5-{4-[3-(4-acetylpiperazine-1-carbonyl)phenyl]quinazolin-6-yl}-2-methoxypyridine-3-carbonitrile, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Discovery and Pharmacological Characterization of Novel Quinazoline-Based PI3K Delta-Selective Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
2Y6R
 
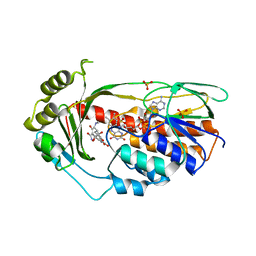 | | Structure of the TetX monooxygenase in complex with the substrate 7- chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Wright, G.D, Hinrichs, W. | | Deposit date: | 2011-01-25 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
3L1V
 
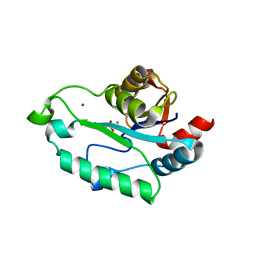 | | Crystal structure of GmhB from E. coli in complex with calcium and phosphate. | | Descriptor: | CALCIUM ION, D,D-heptose 1,7-bisphosphate phosphatase, PHOSPHATE ION, ... | | Authors: | Sugiman-Marangos, S.N, Junop, M.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and kinetic characterization of the LPS biosynthetic enzyme D-alpha,beta-D-heptose-1,7-bisphosphate phosphatase (GmhB) from Escherichia coli.
Biochemistry, 49, 2010
|
|
2Y6Q
 
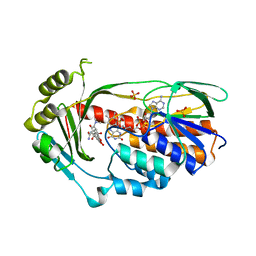 | | Structure of the TetX monooxygenase in complex with the substrate 7- Iodtetracycline | | Descriptor: | 7-IODOTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Wright, G.D, Hinrichs, W. | | Deposit date: | 2011-01-25 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
6DHG
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.5 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
1XW4
 
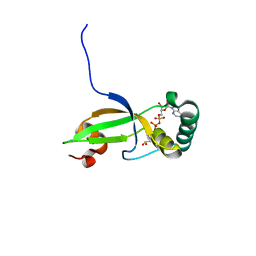 | | Crystal Structure of Human Sulfiredoxin (Srx) in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Sulfiredoxin | | Authors: | Murray, M.S, Jonsson, T.J, Johnson, L.C, Poole, L.B, Lowther, W.T. | | Deposit date: | 2004-10-29 | | Release date: | 2005-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the retroreduction of inactivated peroxiredoxins by human sulfiredoxin.
Biochemistry, 44, 2005
|
|
1XD9
 
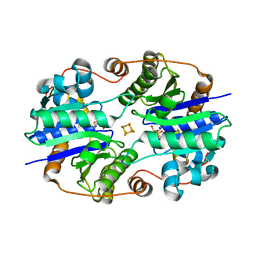 | | Crystal Structure of the Nitrogenase Fe protein Asp39Asn with MgADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Jang, S.B, Jeong, M.S, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical implications of single amino acid substitutions in the nucleotide-dependent switch regions of the nitrogenase Fe protein from Azotobacter vinelandii
J.Biol.Inorg.Chem., 9, 2004
|
|
3KJQ
 
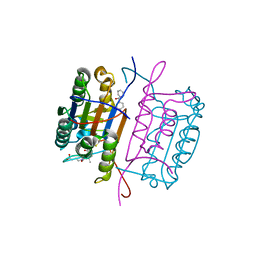 | | Caspase 8 with covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,8R)-2-(3-carboxypropyl)-8-(2-{[(4-chlorophenyl)acetyl]amino}ethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
3MM0
 
 | | Crystal structure of chimeric avidin | | Descriptor: | Avidin, Avidin-related protein 4/5 | | Authors: | Livnah, O, Eisenberg-Domovich, Y, Maatta, J.A.E, Kulomaa, M.S, Hytonen, V.P, Nordlund, H.R. | | Deposit date: | 2010-04-19 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chimeric avidin shows stability against harsh chemical conditions-biochemical analysis and 3D structure.
Biotechnol.Bioeng., 108, 2011
|
|
1XVQ
 
 | | Crystal structure of thiol peroxidase from Mycobacterium tuberculosis | | Descriptor: | AMMONIUM ION, YTTRIUM (III) ION, thiol peroxidase | | Authors: | Rho, B.S, Pedelacq, J.D, Hung, L.W, Holton, J.M, Vigil, D, Kim, S.I, Park, M.S, Terwilliger, T.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-10-28 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and Structural Characterization of a Thiol Peroxidase from Mycobacterium tuberculosis.
J.Mol.Biol., 361, 2006
|
|
1XWR
 
 | | Crystal structure of the coliphage lambda transcription activator protein CII | | Descriptor: | ISOPROPYL ALCOHOL, Regulatory protein CII | | Authors: | Datta, A.B, Panjikar, S, Weiss, M.S, Chakrabarti, P, Parrack, P. | | Deposit date: | 2004-11-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of {lambda} CII: Implications for recognition of direct-repeat DNA by an unusual tetrameric organization
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2YNT
 
 | | GIM-1-3Mol native. Crystal structures of Pseudomonas aeruginosa GIM- 1: active site plasticity in metallo-beta-lactamases | | Descriptor: | GIM-1 PROTEIN, GLYCEROL, ZINC ION | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
1O8W
 
 | | Radiation-reduced Tryparedoxin-I | | Descriptor: | TRYPAREDOXIN | | Authors: | Alphey, M.S, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-12-09 | | Release date: | 2003-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tryparedoxins from Crithidia Fasciculata and Trypanosoma Brucei: Photoreduction of the Redox Disulfide Using Synchrotron Radiation and Evidence for a Conformational Switch Implicated in Function
J.Biol.Chem., 278, 2003
|
|
2YNW
 
 | | GIM-1-2Mol native. Crystal structures of Pseudomonas aeruginosa GIM- 1: active site plasticity in metallo-beta-lactamases | | Descriptor: | GIM-1 PROTEIN, GLYCEROL, SULFATE ION, ... | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
