8UK5
 
 | | Crystal structure of the bromodomain of human ATAD2B in complex with histone H4S1(ph)K5ac | | Descriptor: | ATPase family AAA domain-containing protein 2B, Histone H4S1(ph)K5ac | | Authors: | Montgomery, C, Phillips, M, Nix, J.C, Glass, K.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
J.Med.Chem., 67, 2024
|
|
1FBY
 
 | | CRYSTAL STRUCTURE OF THE HUMAN RXR ALPHA LIGAND BINDING DOMAIN BOUND TO 9-CIS RETINOIC ACID | | Descriptor: | (9cis)-retinoic acid, RETINOIC ACID RECEPTOR RXR-ALPHA | | Authors: | Egea, P.F, Mitschler, A, Rochel, N, Ruff, M, Chambon, P, Moras, D. | | Deposit date: | 2000-07-17 | | Release date: | 2000-07-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid.
EMBO J., 19, 2000
|
|
7UQA
 
 | | Crystal structure of the small Ultra-Red Fluorescent Protein (smURFP) | | Descriptor: | CHLORIDE ION, SODIUM ION, small Ultra-Red Fluorescent Protein (smURFP) | | Authors: | Maiti, A, Buffalo, C.Z, Saurabh, S, Montecinos-Franjola, F, Hachey, J.S, Conlon, W.J, Tran, G.N, Drobizhev, M, Moerner, W.E, Ghosh, P, Matsuo, H, Tsien, R.Y, Lin, J.Y, Rodriguez, E.A. | | Deposit date: | 2022-04-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural and photophysical characterization of the small ultra-red fluorescent protein.
Nat Commun, 14, 2023
|
|
8U2Y
 
 | | Solution structure of the PHD6 finger of MLL4 bound to TET3 | | Descriptor: | Histone-lysine N-methyltransferase 2D, Methylcytosine dioxygenase TET3, ZINC ION | | Authors: | Mohid, S.A, Zandian, M, Zhang, Y, Kutateladze, T.G. | | Deposit date: | 2023-09-06 | | Release date: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | MLL4 binds TET3.
Structure, 32, 2024
|
|
1F5V
 
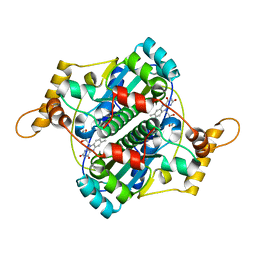 | | STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF A FLAVOPROTEIN FROM ESCHERICHIA COLI THAT REDUCES NITROCOMPOUNDS. ALTERATION OF PYRIDINE NUCLEOTIDE BINDING BY A SINGLE AMINO ACID SUBSTITUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NADPH NITROREDUCTASE | | Authors: | Kobori, T, Sasaki, H, Lee, W.C, Zenno, S, Saigo, K, Murphy, M.E.P, Tanokura, M. | | Deposit date: | 2000-06-17 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution.
J.Biol.Chem., 276, 2001
|
|
8UY1
 
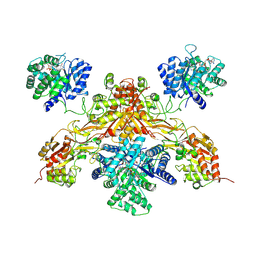 | |
8UF6
 
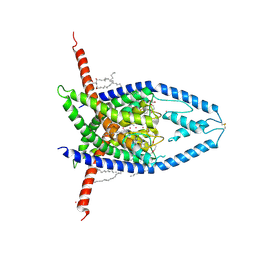 | |
1FHZ
 
 | | PSORALEN CROSS-LINKED D(CCGGTACCGG) FORMS HOLLIDAY JUNCTION | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3') | | Authors: | Eichman, B.F, Mooers, B.H.M, Alberti, M, Hearst, J.E, Ho, P.S. | | Deposit date: | 2000-08-02 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of psoralen cross-linked DNAs: drug-dependent formation of Holliday junctions.
J.Mol.Biol., 308, 2001
|
|
1F62
 
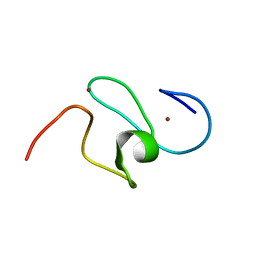 | | WSTF-PHD | | Descriptor: | TRANSCRIPTION FACTOR WSTF, ZINC ION | | Authors: | Pascual, J, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2000-06-19 | | Release date: | 2000-12-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor.
J.Mol.Biol., 304, 2000
|
|
8UY2
 
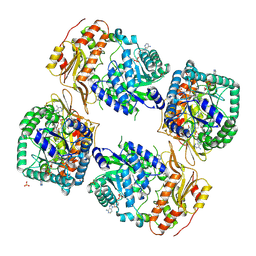 | | Methylenetetrahydrofolate reductase from Chaetomium thermophilum DSM 1495, AdoMet-bound, Inhibited (T) State | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yamada, K, Mendoza, J, Koutmos, M. | | Deposit date: | 2023-11-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural basis of S-adenosylmethionine-dependent allosteric transition from active to inactive states in methylenetetrahydrofolate reductase.
Nat Commun, 15, 2024
|
|
6JZJ
 
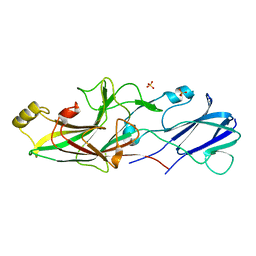 | | Structure of FimA type-2 (FimA2) prepilin of the type V major fimbrium | | Descriptor: | Major fimbrium subunit FimA type-2, SULFATE ION | | Authors: | Okada, K, Shoji, M, Nakayam, K, Imada, K. | | Deposit date: | 2019-05-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
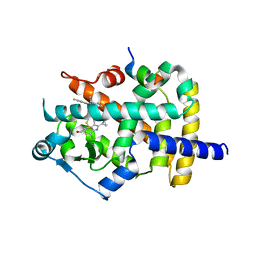
6JZZ
 
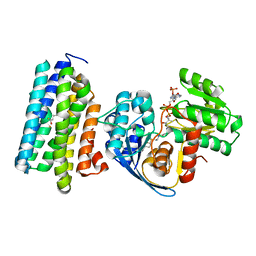 | | The crystal structure of AAR-C294S in complex with ADO. | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6K0T
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-17 | | Descriptor: | 3-[(1~{E})-1-[8-[(8-chloranyl-2-cyclopropyl-imidazo[1,2-a]pyridin-3-yl)methyl]-3-fluoranyl-6~{H}-benzo[c][1]benzoxepin-11-ylidene]ethyl]-4~{H}-1,2,4-oxadiazol-5-one, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Suzuki, M, Yamamoto, K, Takahashi, Y, Saito, J. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Development of a novel class of peroxisome proliferator-activated receptor (PPAR) gamma ligands as an anticancer agent with a unique binding mode based on a non-thiazolidinedione scaffold.
Bioorg.Med.Chem., 27, 2019
|
|
1F6X
 
 | |
8Y0W
 
 | | dormant ribosome with eIF5A, eEF2 and SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
1FEM
 
 | | CRYSTALLOGRAPHIC STUDIES ON COMPLEXES BETWEEN RETINOIDS AND PLASMA RETINOL-BINDING PROTEIN | | Descriptor: | RETINOIC ACID, RETINOL BINDING PROTEIN | | Authors: | Zanotti, G, Marcello, M, Malpeli, G, Sartori, G, Berni, R. | | Deposit date: | 1994-08-29 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies on complexes between retinoids and plasma retinol-binding protein.
J.Biol.Chem., 269, 1994
|
|
1F6T
 
 | | STRUCTURE OF THE NUCLEOSIDE DIPHOSPHATE KINASE/ALPHA-BORANO(RP)-TDP.MG COMPLEX | | Descriptor: | 2*-DEOXY-THYMIDINE-5*-ALPHA BORANO DIPHOSPHATE (ISOMER RP), MAGNESIUM ION, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE) | | Authors: | Guerreiro, C, Boretto, J, Janin, J, Veron, M, Canard, B, Schneider, B, Sarfati, S, Deville-Bonne, D. | | Deposit date: | 2000-06-23 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase.
EMBO J., 19, 2000
|
|
7VF4
 
 | | Crystal structure of Vps75 from Candida albicans | | Descriptor: | CHLORIDE ION, SODIUM ION, Vps75 | | Authors: | Wang, W, Chen, X, Yang, Z, Chen, X, Li, C, Wang, M. | | Deposit date: | 2021-09-10 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of histone chaperone Vps75 from Candida albicans.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
8Y0X
 
 | | Dormant ribosome with SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
8Y4Z
 
 | | Monomeric HERC5 HECT c-lobe structure in solution | | Descriptor: | E3 ISG15--protein ligase HERC5 | | Authors: | Dag, C, Lambert, M, Kahraman, K, Lohn, F, Lee, W, Gocenler, O, Guntert, P, Dotsch, V. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Monomeric HERC5 HECT c-lobe structure in solution
To Be Published
|
|
1F9X
 
 | | AVERAGE NMR SOLUTION STRUCTURE OF THE BIR-3 DOMAIN OF XIAP | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN XIAP, ZINC ION | | Authors: | Sun, C, Cai, M, Meadows, R.P, Fesik, S.W. | | Deposit date: | 2000-07-11 | | Release date: | 2001-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP.
J.Biol.Chem., 275, 2000
|
|
6HTI
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 6 | | Descriptor: | GLYCEROL, Histone deacetylase, POTASSIUM ION, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HTT
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 7 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
1FFO
 
 | | CRYSTAL STRUCTURE OF MURINE CLASS I H-2DB COMPLEXED WITH SYNTHETIC PEPTIDE GP33 (C9M/K1A) | | Descriptor: | BETA-2 MICROGLOBULIN BETA CHAIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B, ... | | Authors: | Wang, B, Sharma, A, Maile, R, Saad, M, Collins, E.J, Frelinger, J.A. | | Deposit date: | 2000-07-25 | | Release date: | 2002-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Peptidic termini play a significant role in TCR recognition.
J.Immunol., 169, 2002
|
|
8Y0U
 
 | | dormant ribosome with STM1 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
