4TT2
 
 | | Crystal structure of ATAD2A bromodomain complexed with H4(1-20)K5Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4K5Ac | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
3D7A
 
 | |
4RKN
 
 | | Wolinella succinogenes octaheme sulfite reductase MccA, form II | | Descriptor: | COPPER (II) ION, DITHIONITE, MccA, ... | | Authors: | Hermann, B, Kern, M, La Pietra, L, Simon, J, Einsle, O. | | Deposit date: | 2014-10-13 | | Release date: | 2015-02-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The octahaem MccA is a haem c-copper sulfite reductase.
Nature, 520, 2015
|
|
4TOF
 
 | | 1.65A resolution structure of BfrB (C89S, K96C) crystal form 1 from Pseudomonas aeruginosa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bacterioferritin, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4TQE
 
 | |
7EY3
 
 | | Double cysteine mutations in T1 lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Hamdan, S.H, Leow, T.C, Yahaya, N.M, Ali, M.S.M, Jonet, M.A, Mohamad Aris, S.N.A, Maiangwa, J. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Knotting terminal ends of mutant T1 lipase with disulfide bond improved structure rigidity and stability.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
4RRP
 
 | | Crystal Structure of the Fab complexed with antigen Asf1p, Northeast Structural Genomics Consortium (NESG) Target PdR16 | | Descriptor: | Antigen Asf1p, DI(HYDROXYETHYL)ETHER, Fab antibody, ... | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Mao, L, Xiao, R, Oconnell, P.T, Maglaqui, M, Bailey, L, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structure of the Fab complexed with antigen Asf1p, Northeast Structural Genomics Consortium (NESG) Target PdR16
To be Published
|
|
3D3Q
 
 | | Crystal structure of tRNA delta(2)-isopentenylpyrophosphate transferase (SE0981) from Staphylococcus epidermidis. Northeast Structural Genomics Consortium target SeR100 | | Descriptor: | tRNA delta(2)-isopentenylpyrophosphate transferase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, L, Xiao, R, Maglaqui, M, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of tRNA delta(2)-isopentenylpyrophosphate transferase (SE0981) from Staphylococcus epidermidis.
To be Published
|
|
7KKF
 
 | |
6YBS
 
 | | Structure of a human 48S translational initiation complex - head | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
4RZ0
 
 | | Crystal Structure of Plasmodium falciparum putative histone methyltransferase PFL0690c | | Descriptor: | PFL0690c, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-17 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Crystal Structure of Plasmodium falciparum putative histone methyltransferase PFL0690c
To be Published
|
|
4S1K
 
 | | Structure of Uranotaenia sapphirina cypovirus (CPV17) polyhedrin at 100 K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Ginn, H.M, Messerschmidt, M, Ji, X, Zhang, H, Axford, D, Gildea, R.J, Winter, G, Brewster, A.S, Hattne, J, Wagner, A, Grimes, J.M, Evans, G, Sauter, N.K, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of CPV17 polyhedrin determined by the improved analysis of serial femtosecond crystallographic data.
Nat Commun, 6, 2015
|
|
4P6S
 
 | | Crystal Structure of tyrosinase from Bacillus megaterium with L-DOPA in the active site | | Descriptor: | 3,4-DIHYDROXYPHENYLALANINE, Tyrosinase, ZINC ION | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2014-03-25 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins.
Nat Commun, 5, 2014
|
|
4S3E
 
 | | IspG in complex with Inhibitor 7 (compound 1061) | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4S1D
 
 | | Structure of IgG1 Fab fragment in complex with Biotincytidinamide | | Descriptor: | MAB M33 FAB FRAGMENT, heavy chain, light chain, ... | | Authors: | Dengl, S, Hoffmann, E, Grote, M, Wagner, C, Mundigl, O, Georges, G, Theorey, I, Stubenrauch, K.-G, Bujotzek, A, Josel, H.-P, Dziadek, S, Benz, J, Brinkmann, U. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hapten-directed spontaneous disulfide shuffling: a universal technology for site-directed covalent coupling of payloads to antibodies.
Faseb J., 29, 2015
|
|
3CZX
 
 | |
4TNF
 
 | |
4TOE
 
 | | 2.20A resolution structure of Iron Bound BfrB (D34F) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin, FE (II) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
7X1N
 
 | | Crystal structure of MEF2D-MRE complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*G)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), Myocyte enhancer factor 2D/deleted in azoospermia associated protein 1 fusion protein | | Authors: | Zhang, H, Zhang, M, Wang, Q.Q, Chen, Z, Chen, S.J, Meng, G. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.315 Å) | | Cite: | Functional, structural, and molecular characterizations of the leukemogenic driver MEF2D-HNRNPUL1 fusion.
Blood, 140, 2022
|
|
3D3N
 
 | | Crystal structure of lipase/esterase (lp_2923) from Lactobacillus plantarum. Northeast Structural Genomics Consortium target LpR108 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Putative lipase/esterase | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Mao, L, Janjua, H, Xiao, R, Ciccosanti, C, Maglaqui, M, Foote, E.L, Zhao, L, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lipase/esterase (lp_2923) from Lactobacillus plantarum. Northeast Structural Genomics Consortium target LpR108
To be Published
|
|
4TU6
 
 | | Crystal structure of apo ATAD2A bromodomain with N1064 alternate conformation | | Descriptor: | ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4RLD
 
 | |
4PI9
 
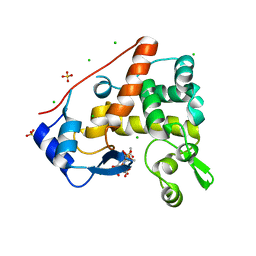 | | Crystal structure of S. Aureus Autolysin E in complex with muropeptide NAM-L-ALA-D-iGLU | | Descriptor: | (4R)-4-[[(2S)-2-[[(2R)-2-[(2R,3S,4R,5R,6R)-5-acetamido-2-(hydroxymethyl)-3,6-bis(oxidanyl)oxan-4-yl]oxypropanoyl]amino]propanoyl]amino]-5-azanyl-5-oxidanylidene-pentanoic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers.
IUCrJ, 4, 2017
|
|
4RQW
 
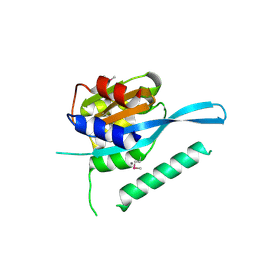 | | Crystal structure of Myc3 N-terminal JAZ-binding domain [44-238] from Arabidopsis | | Descriptor: | CALCIUM ION, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-05 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4RTI
 
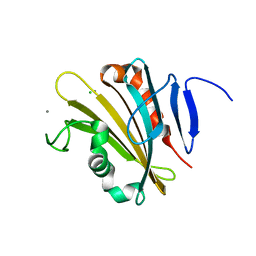 | | The crystal structure of PsbP from Spinacia oleracea | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Oxygen-evolving enhancer protein 2, ... | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
