7OCJ
 
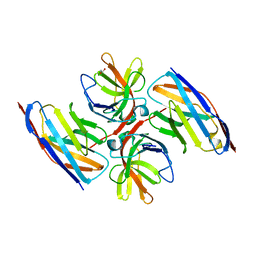 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS2(Nb14509) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, NbSOS2 (14509) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2021-04-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
5YGY
 
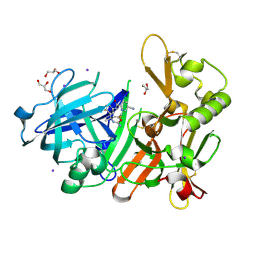 | | Crystal Structure of BACE1 in complex with (S)-N-(3-(2-amino-6-(fluoromethyl)-4 -methyl-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-cyanopicolinamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fuchino, K, Mitsuoka, Y, Masui, M, Kurose, N, Yoshida, S, Komano, K, Yamamoto, T, Ogawa, M, Unemura, C, Hosono, M, Ito, H, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Miyajima, H, Hiroyama, S, Koyabu, K, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design of Novel 1,3-Oxazine Based beta-Secretase (BACE1) Inhibitors: Incorporation of a Double Bond To Reduce P-gp Efflux Leading to Robust A beta Reduction in the Brain
J. Med. Chem., 61, 2018
|
|
7VVI
 
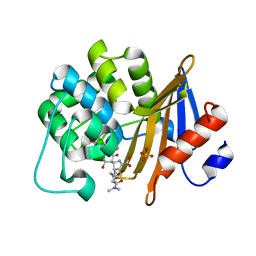 | | OXA-58 crystal structure of acylated meropenem complex | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, SULFATE ION | | Authors: | Saino, H, Sugiyabu, T, Miyano, M. | | Deposit date: | 2021-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | OXA-58 crystal structure of acylated meropenem complex
to be published
|
|
1DIB
 
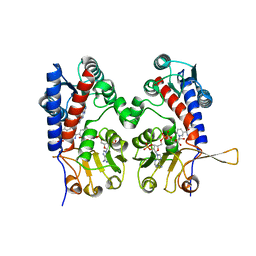 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY345899 | | Descriptor: | 4-(7-AMINO-9-HYDROXY-1-OXO-3,3A,4,5-TETRAHYDRO-2,5,6,8,9B-PENTAAZA-CYCLOPENTA[A]NAPHTHALEN-2-YL)-PHENYLCARBONYL-GLUTAMI C ACID, METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
7OJ5
 
 | | Cryo-EM structure of Medicago truncatula HISN5 protein | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Ruszkowski, M, Witek, W. | | Deposit date: | 2021-05-13 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
1DIG
 
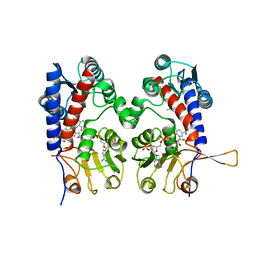 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY374571 | | Descriptor: | ACETATE ION, METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
1QHI
 
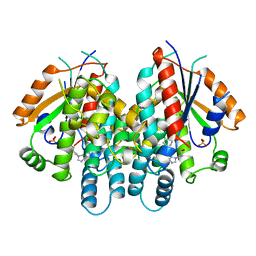 | | HERPES SIMPLEX VIRUS TYPE-I THYMIDINE KINASE COMPLEXED WITH A NOVEL NON-SUBSTRATE INHIBITOR, 9-(4-HYDROXYBUTYL)-N2-PHENYLGUANINE | | Descriptor: | 9-(4-HYDROXYBUTYL)-N2-PHENYLGUANINE, PROTEIN (THYMIDINE KINASE), SULFATE ION | | Authors: | Bennett, M.S, Wien, F, Champness, J.N, Batuwangala, T, Rutherford, T, Summers, W.C, Sun, H, Wright, G, Sanderson, M.R. | | Deposit date: | 1999-05-12 | | Release date: | 1999-07-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure to 1.9 A resolution of a complex with herpes simplex virus type-1 thymidine kinase of a novel, non-substrate inhibitor: X-ray crystallographic comparison with binding of aciclovir.
FEBS Lett., 443, 1999
|
|
1DIA
 
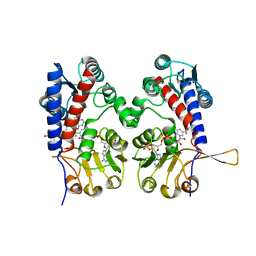 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY249543 | | Descriptor: | METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [[[2-AMINO-5,6,7,8-TETRAHYDRO-4-HYDROXY-PYRIDO[2,3-D]PYRIMIDIN-6-YL]-ETHYL]-PHENYL]-CARBONYL-GLUTAMIC ACID | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
4R50
 
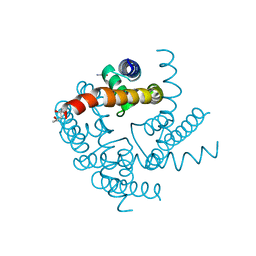 | | Crystal Structure of CNG mimicking NaK-ETPP mutant cocrystallized with Li+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, Potassium channel protein | | Authors: | De March, M, Napolitano, L.M.R, Onesti, S. | | Deposit date: | 2014-08-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7T9U
 
 | | Crystal structure of hSTING with an agonist (SHR169224) | | Descriptor: | (3S,4S)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4-(prop-2-en-1-yl)-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
7T9V
 
 | | Crystal structure of hSTING with the agonist, SHR171032 | | Descriptor: | (3S,4S)-4-(3-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazol-1-yl}propyl)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
8HJ9
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | Descriptor: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HJ3
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | Descriptor: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HHO
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | Descriptor: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HIQ
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | Descriptor: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | Deposit date: | 2022-11-21 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
7OLZ
 
 | | Crystal structure of the SARS-CoV-2 RBD with neutralizing-VHHs Re5D06 and Re9F06 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Nanobody Re5D06, Nanobody Re9F06, ... | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
8HIZ
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | Descriptor: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
7ON5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing nanobody Re5D06 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Nanobody Re5D06 | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
1DKE
 
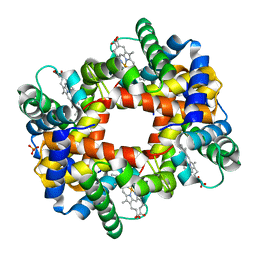 | | NI BETA HEME HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN: ALPHA CHAIN, HEMOGLOBIN: BETA CHAIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bruno, S, Bettatti, S, Mozzarelli, A, Bolognesi, M, Deriu, D, Rosano, C, Tsuneshige, A, Yonetani, T, Henry, E.R. | | Deposit date: | 1999-12-07 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oxygen binding by alpha(Fe2+)2beta(Ni2+)2 hemoglobin crystals.
Protein Sci., 9, 2000
|
|
7W9W
 
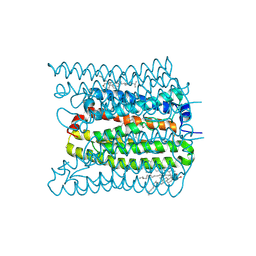 | | 2.02 angstrom cryo-EM structure of the pump-like channelrhodopsin ChRmine | | Descriptor: | CHOLESTEROL, ChRmine, PALMITIC ACID, ... | | Authors: | Kishi, K.E, Kim, Y, Fukuda, M, Yamashita, K, Deisseroth, K, Kato, H.E. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine.
Cell, 185, 2022
|
|
7TAK
 
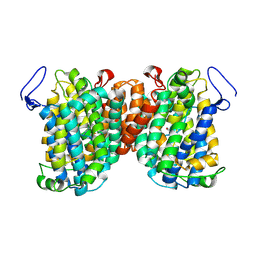 | | Structure of a NAT transporter | | Descriptor: | GUANINE, Putative membrane protein PurT | | Authors: | Weng, J, Zhou, X, Ren, Z, Chen, K, Zhou, M. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.79828 Å) | | Cite: | Insight into the mechanism of H + -coupled nucleobase transport.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7T5O
 
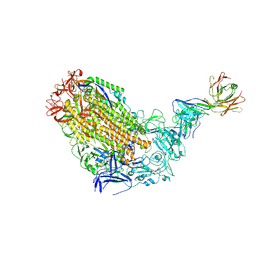 | | VFLIP Spike Trimer with GAR03 | | Descriptor: | GAR03 Fab heavy chain, GAR03 Fab light chain, Spike glycoprotein | | Authors: | Sobti, M, Stewart, A.G, Rouet, R, Langley, D.B. | | Deposit date: | 2021-12-12 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients.
Nat Commun, 14, 2023
|
|
5Y40
 
 | | Structure of the periplasmic domain of the MotB L119P mutant from Salmonella (crystal form 2) | | Descriptor: | Motility protein B | | Authors: | Takao, M, Kojima, S, Sakuma, M, Homma, M, Imada, K. | | Deposit date: | 2017-07-31 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Helix Rearrangement in the Periplasmic Domain of the Flagellar Stator B Subunit Activates Peptidoglycan Binding and Ion Influx.
Structure, 26, 2018
|
|
8K8E
 
 | | Human gamma-secretase in complex with a substrate mimetic | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shi, Y.G, Zhou, R, Wolfe, M.S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Familial Alzheimer mutations stabilize synaptotoxic gamma-secretase-substrate complexes.
Cell Rep, 43, 2024
|
|
1DK0
 
 | | CRYSTAL STRUCTURE OF THE HEMOPHORE HASA FROM SERRATIA MARCESCENS CRYSTAL FORM P2(1), PH8 | | Descriptor: | HEME-BINDING PROTEIN A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnoux, P, Haser, R, Izadi-Pruneyre, N, Lecroisey, A, Czjzek, M. | | Deposit date: | 1999-12-06 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Functional aspects of the heme bound hemophore HasA by structural analysis of various crystal forms.
Proteins, 41, 2000
|
|
