4YSA
 
 | | Completely oxidized structure of copper nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | COPPER (II) ION, Nitrite reductase, SODIUM ION | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YUM
 
 | | Multiconformer synchrotron model of CypA at 300 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
1ZK6
 
 | | NMR solution structure of B. subtilis PrsA PPIase | | Descriptor: | Foldase protein prsA | | Authors: | Tossavainen, H, Permi, P, Purhonen, S.L, Sarvas, M, Kilpelainen, I, Seppala, R. | | Deposit date: | 2005-05-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and characterization of substrate binding site of the PPIase domain of PrsA protein from Bacillus subtilis
Febs Lett., 580, 2006
|
|
2X7J
 
 | | Structure of the menaquinone biosynthesis protein MenD from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, 2-SUCCINYL-5-ENOLPYRUVYL-6-HYDROXY-3-CYCLOHEXENE -1-CARBOXYLATE SYNTHASE, MANGANESE (II) ION, ... | | Authors: | Dawson, A, Chen, M, Fyfe, P.K, Guo, Z, Hunter, W.N. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Reactivity of Bacillus Subtilis Mend Catalyzing the First Committed Step in Menaquinone Biosynthesis.
J.Mol.Biol., 401, 2010
|
|
2WTY
 
 | | Crystal structure of the homodimeric MafB in complex with the T-MARE binding site | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP *GP*CP*AP*AP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*TP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP *GP*CP*AP*AP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Consani Textor, L, Holton, S, Wilmanns, M. | | Deposit date: | 2009-09-25 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of a bZIP Transcription Factor with Homo/Heterodimer-Induced DNA-Binding Preference.
Structure, 22, 2014
|
|
3NC0
 
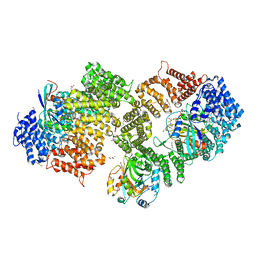 | | Crystal structure of the HIV-1 Rev NES-CRM1-RanGTP nuclear export complex (crystal II) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Exportin-1, GLYCEROL, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2WX0
 
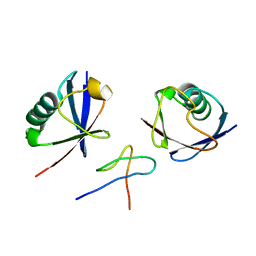 | | TAB2 NZF DOMAIN IN COMPLEX WITH Lys63-linked di-ubiquitin, P21 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 2, UBIQUITIN, ZINC ION | | Authors: | Kulathu, Y, Akutsu, M, Bremm, A, Hofmann, K, Komander, D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two-Sided Ubiquitin Binding Explains Specificity of the Tab2 Nzf Domain
Nat.Struct.Mol.Biol., 16, 2009
|
|
8CH0
 
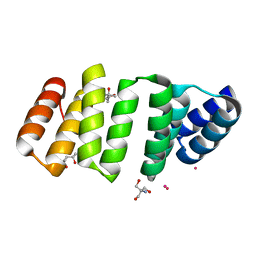 | | Crystal structure of an 8-repeat consensus TPR superhelix with Gadolinium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Consensus tetratricopeptide repeat protein, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
2WOC
 
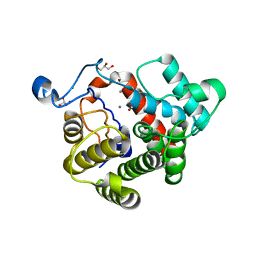 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1ZP0
 
 | | Crystal Structure of Mitochondrial Respiratory Complex II bound with 3-nitropropionate and 2-thenoyltrifluoroacetone | | Descriptor: | 3-NITROPROPANOIC ACID, 4,4,4-TRIFLUORO-1-THIEN-2-YLBUTANE-1,3-DIONE, FAD-binding protein, ... | | Authors: | Sun, F, Huo, X, Zhai, Y, Wang, A, Xu, J, Su, D, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-16 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Mitochondrial Respiratory Membrane Protein Complex II
Cell(Cambridge,Mass.), 121, 2005
|
|
8C0B
 
 | | CryoEM structure of Aspergillus nidulans UTP-glucose-1-phosphate uridylyltransferase | | Descriptor: | UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Han, X, D Angelo, C, Otamendi, A, Cifuente, J.O, de Astigarraga, E, Ochoa-Lizarralde, B, Grininger, M, Routier, F.H, Guerin, M.E, Fuehring, J, Etxebeste, O, Connell, S.R. | | Deposit date: | 2022-12-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | CryoEM analysis of the essential native UDP-glucose pyrophosphorylase from Aspergillus nidulans reveals key conformations for activity regulation and function.
Mbio, 14, 2023
|
|
1ZFE
 
 | | GCA Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*GP*CP*AP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
8CIG
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix in tris Buffer with Calcium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8BZO
 
 | | Cryo-EM structure of CDK2-CyclinA in complex with p27 from the SCFSKP2 E3 ligase Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-15 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
5CZU
 
 | | Crystal structure of FeCat-Fn | | Descriptor: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | Authors: | Abe, S, Nakajima, H, Kondo, M, Nakane, T, Nakao, T, Ueno, T, Watanabe, Y. | | Deposit date: | 2015-08-01 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Construction of an enterobactin analogue with symmetrically arranged monomer subunits of ferritin
Chem.Commun.(Camb.), 51, 2015
|
|
8BYA
 
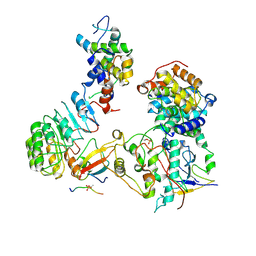 | | Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
8C50
 
 | |
5D0C
 
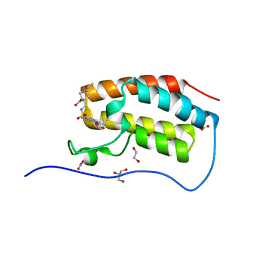 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-08-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
1YXJ
 
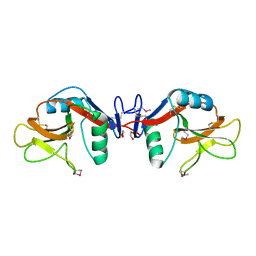 | | Crystal structure of human lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) at low pH | | Descriptor: | 1,2-ETHANEDIOL, oxidised low density lipoprotein (lectin-like) receptor 1 | | Authors: | Ohki, I, Ishigaki, T, Oyama, T, Matsunaga, S, Xie, Q, Ohnishi-Kameyama, M, Murata, T, Tsuchiya, D, Machida, S, Morikawa, K, Tate, S. | | Deposit date: | 2005-02-22 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human lectin-like, oxidized low-density lipoprotein receptor 1 ligand binding domain and its ligand recognition mode to OxLDL.
Structure, 13, 2005
|
|
8BYL
 
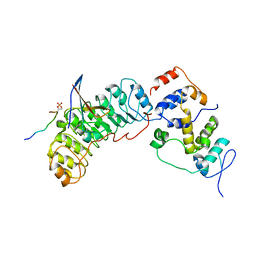 | | Cryo-EM structure of SKP1-SKP2-CKS1 from the SCFSKP2 E3 ligase complex | | Descriptor: | Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, S-phase kinase-associated protein 1, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
3N1N
 
 | | Crystal structure of the complex of type I ribosome inactivating protein with guanine at 2.2A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANINE, Ribosome inactivating protein | | Authors: | Kushwaha, G.S, Singh, N, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-16 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states
Biochim.Biophys.Acta, 1824, 2012
|
|
1ZHV
 
 | | X-ray Crystal Structure Protein Atu0741 from Agobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR8. | | Descriptor: | hypothetical protein Atu0741 | | Authors: | Vorobiev, S.M, Kuzin, A, Abashidze, M, Forouhar, F, Xio, R, Ma, L.-C, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-26 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of hypothetical protein Atu071 from Agobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR8.
To be Published
|
|
2X49
 
 | | Crystal structure of the C-terminal domain of InvA | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, INVASION PROTEIN INVA, ... | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the C-Terminal Domain of the Salmonella Type III Secretion System Export Apparatus Protein Inva.
Protein Sci., 19, 2010
|
|
4IGY
 
 | | Crystal structure of kirola (Act d 11) - triclinic form | | Descriptor: | CHLORIDE ION, Kirola, UNKNOWN LIGAND | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
8CIH
 
 | | Structure of FL CINP | | Descriptor: | Cyclin-dependent kinase 2-interacting protein, SODIUM ION | | Authors: | Foglizzo, M, Zeqiraj, E. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity.
Cell, 187, 2024
|
|
