8DL7
 
 | | Cryo-EM structure of human ferroportin/slc40 bound to minihepcidin PR73 in nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 11F9 heavy-chain, 11F9 light-chain, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
8EAI
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 16-mer oligo-dT. Class 2 | | Descriptor: | 16-mer oligo-dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8DL8
 
 | | Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
6EO4
 
 | | Physcomitrella patens BBE-like 1 wild-type | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PpBBE-like 1 | | Authors: | Toplak, M, Winkler, A, Macheroux, P. | | Deposit date: | 2017-10-09 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The single berberine bridge enzyme homolog of Physcomitrella patens is a cellobiose oxidase.
FEBS J., 285, 2018
|
|
8EAK
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 46-mer DNA strand. Class 2 | | Descriptor: | 46-mer DNA, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAM
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and DNA. Class 2. Merged particles from datasets with 3 different DNA entities | | Descriptor: | DNA, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAJ
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 46-mer DNA strand. Class 1 | | Descriptor: | 46-mer DNA strand, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAH
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 16-mer oligo-dT. Class 1 | | Descriptor: | 16-mer oligo-dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAF
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 12-mer oligo-dT. Class 1 | | Descriptor: | 12-mer oligo dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
8EAG
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 12-mer oligo-dT. Class 2 | | Descriptor: | 12-mer oligo-dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
5OQP
 
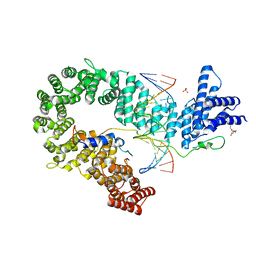 | | Crystal structure of the S. cerevisiae condensin Ycg1-Brn1 subcomplex bound to DNA (crystal form I) | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, DNA (5'-D(*GP*AP*TP*GP*TP*GP*TP*AP*GP*CP*TP*AP*CP*AP*CP*AP*TP*C)-3'), ... | | Authors: | Kschonsak, M, Hassler, M, Haering, C.H. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes.
Cell, 171, 2017
|
|
5OQR
 
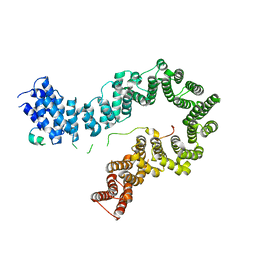 | |
5OSN
 
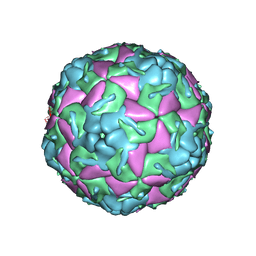 | | Crystal Structure of Bovine Enterovirus 2 determined with Serial Femtosecond X-ray Crystallography | | Descriptor: | Capsid protein, GLUTAMIC ACID, POTASSIUM ION, ... | | Authors: | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Kotecha, A, Kelly, J, Rowlands, D.J, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
4WNX
 
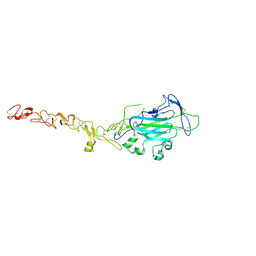 | | Netrin 4 lacking the C-terminal Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McDougall, M, Patel, T, Reuten, R, Meier, M, Koch, M, Stetefeld, J. | | Deposit date: | 2014-10-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes.
Nat Commun, 7, 2016
|
|
4RMI
 
 | | Human Sirt2 in complex with SirReal1 and Ac-Lys-OTC peptide | | Descriptor: | Ac-Lys-OTC peptide, N-(5-benzyl-1,3-thiazol-2-yl)-2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
8BOC
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 19 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-[3,5-bis(trifluoromethyl)phenyl]-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
4RMJ
 
 | | Human Sirt2 in complex with ADP ribose and nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
8VYE
 
 | | SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, S2L20 Light Chain, ... | | Authors: | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-02-08 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8VYG
 
 | | SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Heavy Chain, S309 Light Chain, ... | | Authors: | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-02-08 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8VYF
 
 | | SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, ... | | Authors: | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-02-08 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
4RMH
 
 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4UIB
 
 | | Crystal structure of 3p in complex with tafCPB | | Descriptor: | (2S)-6-AMINO-2-[[(1R)-1-(CYCLOHEXYLMETHYL)-2-OXO-2-[[(2S)-1,7,7-TRIMETHYLNORBORNAN-2-YL]AMINO]ETHYL]ARBAMOYLAMINO]HEXANOIC ACID, ACETATE ION, CARBOXYPEPTIDASE B, ... | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
4UIA
 
 | | Crystal structure of 3a in complex with tafCPB | | Descriptor: | (2S)-6-azanyl-2-[[(2R)-3-cyclohexyl-1-(3-methylbutylamino)-1-oxidanylidene-propan-2-yl]carbamoylamino]hexanoic acid, CARBOXYPEPTIDASE B, ZINC ION | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
4USP
 
 | | X-ray structure of the dimeric CCL2 lectin in native form | | Descriptor: | CCL2 LECTIN, CHLORIDE ION, PHOSPHATE ION | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
5LUR
 
 | | An avidin mutant | | Descriptor: | Avidin, CHLORIDE ION, PROGESTERONE | | Authors: | Agrawal, N, Lehtonen, S.I, Riihimaki, T.A, Kulomaa, M.S, Hytonen, V.P, Johnson, M.S, Airenne, T.T. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An avidin mutant
TO BE PUBLISHED
|
|
